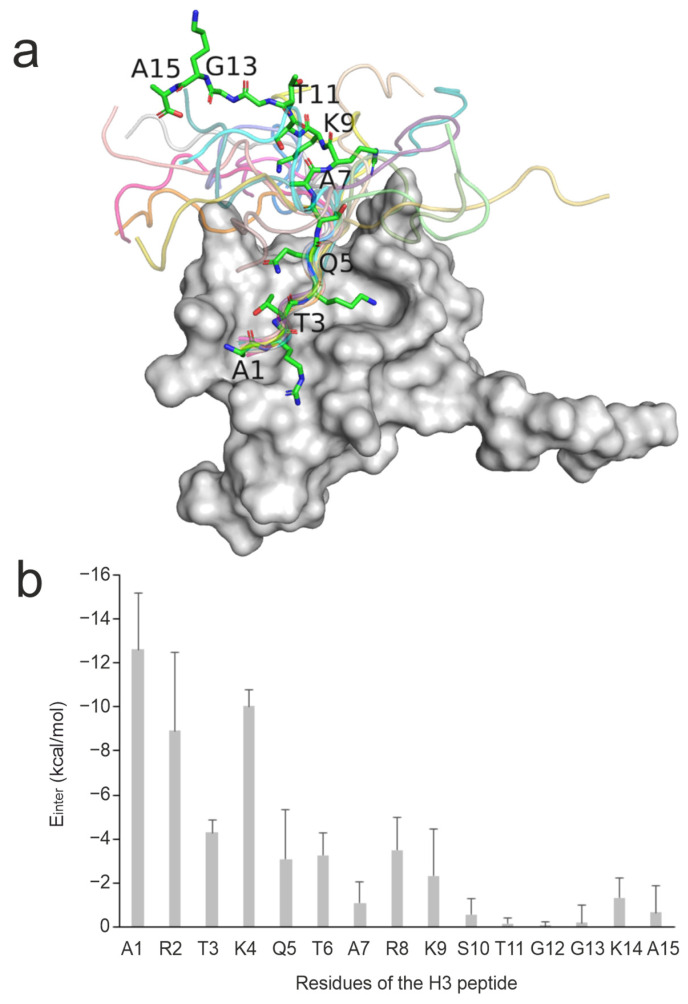
Figure 1.
Structural and per-residue energetic analysis of histone H3 peptide bound to reader protein. (a) NMR solution structure of the BPTF PHD finger domain (grey surface, PDB ID 2fui) in complex with a histone H3 peptide in which the peptide structure in the first model is shown in sticks representation while the remaining models are shown in cartoon representation. (b) The mean (columns) and standard deviations (error bars) of Einter values of all 20 NMR models calculated for the peptide respective residues upon a brief energy minimization.