Abstract
The drug-resistant temporal lobe epilepsy (TLE) has recently been associated with single nucleotide variants (SNVs) in microRNA(miR)-146a (MIR-146A) (rs2910164) and Sodium Voltage-Gated Channel Alpha Subunit 1 (SCN1A) (rs2298771 and rs3812718) genes. Moreover, no studies have shown an association between these SNVs and susceptibility to drug-resistant and drug-responsive TLE in Brazil. Thus, deoxyribonucleic acid (DNA) samples from 120 patients with TLE (55 drug-responsive and 65 drug-resistant) were evaluated by real-time polymerase chain reaction (RT-PCR). A total of 1171 healthy blood donor individuals from the Online Archive of Brazilian Mutations (ABraOM, from Portuguese Arquivo Brasileiro On-line de Mutações), a repository containing genomic variants of the Brazilian population, were added as a control population for the studied SNVs. MIR-146A and SCN1A relative expression was performed by quantitative RT-PCR (qRT-PCR). The statistical analysis protocol was performed using an alpha error of 0.05. TLE patient samples and ABraOM control samples were in Hardy–Weinberg equilibrium for all studied SNVs. For rs2910164, the frequencies of the homozygous genotype (CC) (15.00% vs. 9.65%) and C allele (37.80% vs. 29.97%) were superior in patients with TLE compared to controls with a higher risk for TLE disease [odds ratio (OR) = 1.89 (95% confidence interval (95%CI) = 1.06–3.37); OR = 1.38 (95%CI = 1.04–1.82), respectively]. Drug-responsive patients also presented higher frequencies of the CC genotype [21.81% vs. 9.65%; OR = 2.58 (95%CI = 1.25–5.30)] and C allele [39.09% vs. 29.97%; OR = 1.50 (95%CI = 1.01–2.22)] compared to controls. For rs2298771, the frequency of the heterozygous genotype (AG) (51.67% vs. 40.40%) was superior in patients with TLE compared to controls with a higher risk for TLE disease [OR = 2.42 (95%CI = 1.08–5.41)]. Drug-resistant patients presented a higher AG frequency [56.92% vs. 40.40%; OR = 3.36 (95%CI = 1.04–17.30)] compared to the control group. For rs3812718, the prevalence of genotypes and alleles were similar in both studied groups. The MIR-146A relative expression level was lower in drug-resistant compared to drug-responsive patients for GC (1.6 vs. 0.1, p-value = 0.049) and CC (1.8 vs. 0.6, p-value = 0.039). Also, the SCN1A relative expression levels in samples from TLE patients were significantly higher in AG [2.09 vs. 1.10, p-value = 0.038] and GG (3.19 vs. 1.10, p-value < 0.001) compared to the AA genotype. In conclusion, the rs2910164-CC and rs2298771-AG genotypes are exerting significant risk influence, respectively, on responsive disease and resistant disease, probably due to an upregulated nuclear factor kappa B (NF-kB) and SCN1A loss of function.
Keywords: drug-resistant, drug-responsive, MIR-146a gene, SCN1A gene, single nucleotide variant, temporal lobe epilepsy
1. Introduction
Epilepsy, a chronic disease of the central nervous system, affects individuals of all ages [1] with an average world incidence of around 5.4 per 1000 individuals and an average world lifetime incidence of approximately 7.0 per 1000 individuals [1,2]. The World Health Organization estimates that 50 million people are diagnosed with epilepsy in the world, with 4 million in Brazil [3,4]. Still, about 66% of the epilepsy cases are temporal lobe epilepsy (TLE), classified as focal epilepsy [5]. It is the most common epileptic syndrome in adults, of which 40% are drug-resistant seizures [6].
More than half of epilepsy cases present a genetic basis and a complex inheritance pattern [7,8]. The single nucleotide variants (SNVs) alter amino acids of protein-coding genes and can influence protein function and play a vital role in the pathophysiology of diseases such as epilepsy [9]. SNVs at microRNA (miR)-146a (MIR-146A) and voltage-gated sodium channel SCN1A (Sodium Voltage-Gated Channel Alpha Subunit 1) have been recently related to TLE [10,11,12].
MiRs are small non-coding molecules that bind to messenger RNA (mRNA) and prevent its translation, and recent studies have observed miRs related to epilepsy [12,13]. miR-146a is upregulated in human astrocytes in epileptogenic tissues and it regulates the inflammatory process through nuclear factor-kappa B (NF-kB) signaling [14,15]. The SNV n.60G>C (rs2910164) located at the mature sequence of miR-146a has been studied in Italian, Chinese, and Brazilian populations with contradictory results [11,16,17,18]. In addition, only one Chinese and one Brazilian study have evaluated patients with TLE [11,18].
Epileptic seizure susceptibility versus multidrug resistance has recently been related to the different genotypes from SNVs in the SCN1A gene, including SNV c.3184A>G (rs2298771) and SNV IVS5N+5G>A (rs3812718) [19,20,21,22,23,24,25,26,27,28]. There is only one study that has evaluated the SNV rs3812718 and risk in relation to non-multidrug resistance in patients with TLE [29].
Drug-resistant epilepsy is defined as a failure of adequate trials of two tolerated, appropriately chosen, and used antiepileptic drug schedules (whether as monotherapies or in combination) to achieve sustained seizure freedom [30]. Thus, the main goal of the preliminary study was to show for the first time the association of SNV rs2910164 in the MIR-146A gene, SNVs rs2298771 and rs3812718 in the SCN1A gene, and the susceptibility to drug-resistant and drug-responsive TLE in a Brazilian cohort sample.
2. Results
2.1. Patients with Temporal Lobe Epilepsy and Healthy Controls
This study’s groups comprised 120 patients with TLE, 55 patients diagnosed as drug-responsive (22 males; 33 females; mean age: 45.23 years), and 65 patients diagnosed as drug-resistant (27 males; 38 females; mean age: 51.03 years) (Table 1). The average seizure onset age was earlier in the drug-resistant than in the drug-responsive group (11.09 vs. 23.81 years; p-value = 0.004). The average of Caucasian patients was higher than 70% in both TLE groups (p-value = 0.001). The sides of epileptiform paroxysms were similar in both TLE groups (p-value = 0.066) (Table 1). However, it was observed that brain tissue injury was more significant in the drug-resistant than in the drug-responsive group (90.80% vs. 40.70%; p-value < 0.001) (Table 1). The drug-resistant group used anti-epileptic drug polytherapy in 96.90% of the cases, while the drug-responsive group used monotherapy in 72.20% of the cases (p-value < 0.001) (Table 1).
Table 1.
Clinical variables of the patients with temporal lobe epilepsy enrolled in this study.
| Markers | Groups | Patients |
p-Value (p-Corrected) |
|
|---|---|---|---|---|
| Drug-Resistant n (%) | Drug-Responsive n (%) | |||
| Sex | Female | 38 (58.50) | 33 (60.00) | 1.000 (1.000) a |
| Male | 27 (41.50) | 22 (40.00) | ||
| Age (years) * | 53.50 (49.51–56.00) | 45.00 (33.00–39.00) | 0.118 (0.236) c | |
| Race | White people | 51 (78.50) | 39 (70.90) | 0.001 (0.002) b |
| Pardos (Mixed race) | 13 (20.00) | 4 (7.30) | ||
| Black people | 1 (1.50) | 10 (18.20) | ||
| Asian individuals | 0 (0.00) | 2 (3.60) | ||
| Age of onset (years) * | 10.50 (7.00–13.00) | 15.00 (12.00–19.00) | 0.004 (0.008) c | |
| Electroencephalogram | Not specified | 2 (3.10) | 5 (9.10) | 0.066 (0.132) b |
| Normal | 0 (0.00) | 5 (9.10) | ||
| Bilateral temporal | 12 (18.50) | 8 (14.50) | ||
| Right temporal | 22 (33.80) | 18 (32.70) | ||
| Left temporal | 29 (44.60) | 19 (34.50) | ||
| Structural brain lessions | Yes | 59 (90.80) | 22 (40.70) | <0.001 (<0.001) a |
| No | 6 (9.20) | 32 (59.30) | ||
| Therapy with antiepileptic drug | Monotherapy | 2 (3.10) | 39 (72.20) | <0.001 (<0.001) b |
| Polytherapy | 63 (96.90) | 15 (27.80) | ||
a, Chi-square test; b, Fisher’s exact test; c, Mann–Whitney test; *, median [95% confidence interval (95%CI)]; %, percentage; n, number of individuals. p-corrected was adjusted using Bonferroni correction for multiple comparisons.
2.2. Distribuion of the Genotype and Allele Frequencies for Single Nucleotide Variants (SNVs) rs2910164, rs2298771, and rs3812718 Using Online Archive of Brazilian Mutations (ABraOM, from Portuguese Arquivo Brasileiro On-Line de Mutações) Controls
TLE patient samples and Online Archive of Brazilian Mutations (ABraOM, from Portuguese Arquivo Brasileiro On-line de Mutações) control samples were in Hardy–Weinberg equilibrium for all studied SNVs (Table 2, Table 3 and Table 4).
Table 2.
Comparative association of the variant rs2910164 in microRNA (miR)-146a (MIR-146A) gene and susceptibility to temporal lobe epilepsy and Online Archive of Brazilian Mutations (ABraOM, from Portuguese Arquivo Brasileiro On-line de Mutações) controls.
| Genotypes and Alleles | Patients n (%) a | Controls n (%) b | p-Value (p-Corrected) | OR (95%CI) |
|---|---|---|---|---|
| GG | 49 (40.83) | 582 (49.70) | Reference | |
| GC | 53 (44.17) | 476 (40.65) | 0.213 * (1.000) | 1.32 (0.88–1.99) |
| CC | 18 (15.00) | 113 (9.65) | 0.043 * (1.000) | 1.89 (1.06–3.37) |
| GG + CC | 67 (55.83) | 695 (59.35) | Reference | |
| GC | 53 (44.17) | 476 (40.65) | 0.517 * (1.000) | 1.16 (0.79–1.69) |
| GG + GC | 102 (85.00) | 1058 (90.35) | Reference | |
| CC | 18 (15.00) | 113 (9.65) | 0.090 * (1.000) | 1.65 (0.37–2.83) |
| GC + CC | 71 (59.17) | 589 (50.30) | Reference | |
| GG | 49 (40.83) | 582 (49.70) | 0.079 * (1.000) | 0.70 (0.48–1.02) |
| Allele C | 89 (37.08) | 702 (29.97) | 0.028 * (0.672) | 1.38 (1.04–1.82) |
| Allele G | 151 (62.92) | 1640 (70.03) | Reference | |
| Genotypes and Alleles | Drug-Resistant n (%) c | Controls n (%) | p-Value (p-Corrected) | OR (95%CI) |
| GG | 25 (38.46) | 582 (49.70) | Reference | |
| GC | 34 (52.31) | 476 (40.65) | 0.078 * (1.000) | 1.66 (0.98–2.83) |
| CC | 6 (9.23) | 113 (9.65) | 0.836 * (1.000) | 1.24 (0.50–3.08) |
| GG + CC | 31 (47.69) | 695 (59.35) | Reference | |
| GC | 34 (52.31) | 476 (40.65) | 0.084 * (1.000) | 1.60 (0.97–2.64) |
| GG + GC | 59 (90.77) | 1058 (90.35) | Reference | |
| CC | 6 (9.23) | 113 (9.65) | 0.917 * (1.000) | 0.95 (0.40–2.25) |
| GC + CC | 40 (61.54) | 589 (50.30) | Reference | |
| GG | 25 (38.46) | 582 (49.70) | 0.102 * (1.000) | 0.63 (0.38–1.06) |
| Allele C | 46 (35.38) | 702 (29.97) | 0.227 * (1.000) | 1.28 (0.88–1.85) |
| Allele G | 84 (64.62) | 1640 (70.03) | Reference | |
| Genotypes and Alleles | Drug-Responsive n (%) d | Controls n (%) | p-Value (p-Corrected) | OR (95%CI) |
| GG | 24 (43.63) | 582 (49.70) | Reference | |
| GC | 19 (34.55) | 476 (40.65) | 0.958 * (1.000) | 0.97 (0.52–1.79) |
| CC | 12 (21.82) | 113 (9.65) | 0.015 * (0.360) | 2.58 (1.25–5.30) |
| GG + CC | 36 (65.45) | 695 (59.35) | Reference | |
| GC | 19 (34.55) | 476 (40.65) | 0.447 * (1.000) | 0.77 (0.44–1.36) |
| GG + GC | 43 (78.19) | 1058 (90.35) | Reference | |
| CC | 12 (21.81) | 113 (9.65) | 0.007 * (0.168) | 2.61 (1.34–5.10) |
| GC + CC | 31 (56.36) | 589 (50.30) | Reference | |
| GG | 24 (43.64) | 582 (49.70) | 0.459 * (1.000) | 0.78 (0.45–1.35) |
| Allele C | 43 (39.09) | 702 (29.97) | 0.050 * (1.000) | 1.50 (1.01–2.22) |
| Allele G | 67 (60.91) | 1640 (70.03) | Reference | |
| Genotypes and Alleles | Drug-Resistant n (%) c | Drug-Responsive n (%) d | p-Value (p-Corrected) | OR (95%CI) |
| GG | 25 (38.46) | 24 (43.63) | Reference | |
| GC | 34 (52.31) | 19 (34.55) | 0.255 * (1.000) | 1.72 (0.78–3.80) |
| CC | 6 (9.23) | 12 (21.82) | 0.314 * (1.000) | 0.480 (0.16–1.48) |
| GG + CC | 31 (47.69) | 36 (65.45) | Reference | |
| GC | 34 (52.31) | 19 (34.55) | 0.077 * (1.000) | 2.08 (0.99–4.35) |
| GG + GC | 59 (90.77) | 43 (78.19) | Reference | |
| CC | 6 (9.23) | 12 (21.81) | 0.095 * (1.000) | 0.36 (0.13–1.05) |
| GC + CC | 40 (61.54) | 31 (56.36) | Reference | |
| GG | 25 (38.46) | 24 (43.64) | 0.698 * (1.000) | 0.81 (0.39–1.68) |
| Allele C | 46 (35.38) | 43 (39.09) | 0.647 * (1.000) | 0.85 (0.51–1.44) |
| Allele G | 84 (64.62) | 67 (60.91) | Reference |
*, Chi-square with Yates correction; %, percentage; 95%CI, 95% confidence interval; n, number of individuals; OR, odds ratio. p-value for Hardy–Weinberg equilibrium: (a) 0.842; (b) 0.555; (c) 0.510; (d) 0.126. Values below 0.05 indicate that the sample is out of Hardy–Weinberg equilibrium. Bold type indicates the presence of statistically significant differences in the proportions of genotypes or alleles between the groups of patients and controls. An alpha error of 0.05 was adopted in the statistical analysis. p-corrected was adjusted using Bonferroni correction for multiple comparisons.
Table 3.
Comparative association of the variant rs2298771 in Sodium Voltage-Gated Channel Alpha Subunit 1 (SCN1A) gene and susceptibility to temporal lobe epilepsy and Online Archive of Brazilian Mutations (ABraOM, from Portuguese Arquivo Brasileiro On-line de Mutações) controls.
| Genotypes and Alleles | Patients n (%) a | Controls n (%) b | p-Value (p-Corrected) | OR (95%CI) |
|---|---|---|---|---|
| AA | 51 (42.50) | 569 (48.60) | 0.298 * (1.000) | 1.65 (0.73–3.72) |
| AG | 62 (51.67) | 473 (40.40) | 0.027 * (0.648) | 2.42 (1.08–5.41) |
| GG | 7 (5.83) | 129 (11.00) | Reference | |
| AA + GG | 58 (48.33) | 698 (59.60) | Reference | |
| AG | 62 (51.67) | 473 (40.40) | 0.022 * (0.528) | 1.58 (1.08–2.30) |
| AA + AG | 113 (94.17) | 1042 (89.00) | Reference | |
| GG | 7 (5.83) | 129 (11.00) | 0.109 * (1.000) | 0.50 (0.23–1.10) |
| AG + GG | 69 (57.50) | 602 (51.40) | Reference | |
| AA | 51 (42.50) | 569 (48.60) | 0.240 * (1.000) | 0.78 (0.54–1.14) |
| Allele G | 76 (31.70) | 731 (68.79) | Reference | |
| Allele A | 164 (68.30) | 1611 (31.31) | 0.943 * (1.000) | 0.98 (0.74–1.30) |
| Genotypes and Alleles | Drug-Resistant n (%) c | Controls n (%) | p-Value (p-Corrected) | OR (95%CI) |
| AA | 25 (38.46) | 569 (48.60) | 0.438 ** (1.000) | 1.89 (0.56–9.92) |
| AG | 37 (56.92) | 473 (40.40) | 0.041 ** (0.984) | 3.36 (1.04–17.30) |
| GG | 3 (4.62) | 129 (11.00) | Reference | |
| AA + GG | 28 (43.08) | 698 (59.60) | Reference | |
| AG | 37 (56.92) | 473 (40.40) | 0.012 * (0.288) | 1.95 (1.18–3.23) |
| AA + AG | 62 (95.38) | 1042 (89.00) | Reference | |
| GG | 3 (4.62) | 129 (11.00) | 0.136 ** (1.000) | 0.39 (0.08–1.23) |
| AG + GG | 40 (61.54) | 602 (51.40) | Reference | |
| AA | 25 (38.46) | 569 (48.60) | 0.143 * (1.000) | 0.66 (0.40–1.10) |
| Allele G | 43 (33.10) | 731 (68.79) | Reference | |
| Allele A | 87 (66.90) | 1611 (31.31) | 0.727 * (1.000) | 0.92 (0.63–1.34) |
| Genotypes and Alleles | Drug-Responsive n (%) d | Controls n (%) | p-Value (p-Corrected) | OR (95%CI) |
| AA | 26 (47.27) | 569 (48.60) | 0.665 ** (1.000) | 1.47 (0.50–5.91) |
| AG | 25 (45.46) | 473 (40.40) | 0.464 ** (1.000) | 1.70 (0.57–6.86) |
| GG | 4 (7.27) | 129 (11.00) | Reference | |
| AA + GG | 30 (54.55) | 698 (59.60) | Reference | |
| AG | 25 (45.45) | 473 (40.40) | 0.455 * (1.000) | 1.23 (0.71–2.12) |
| AA + AG | 51 (92.72) | 1042 (89.00) | Reference | |
| GG | 4 (7.28) | 129 (11.00) | 0.537 ** (1.000) | 0.63 (0.16–1.77) |
| AG + GG | 29 (52.72) | 602 (51.40) | Reference | |
| AA | 26 (47.28) | 569 (48.60) | 0.958 * (1.000) | 0.95 (0.55–1.63) |
| Allele G | 33 (33.00) | 731 (68.79) | Reference | |
| Allele A | 77 (77.00) | 1611 (31.31) | 0.871 * (1.000) | 1.06 (0.70–1.61) |
| Genotypes and Alleles | Drug-Resistant n (%) c | Drug-Responsive n (%) d | p-Value (p-Corrected) | OR (95%CI) |
| AA | 25 (38.46) | 26 (47.27) | 1.000 ** (1.000) | 1.28 (0.19–9.61) |
| AG | 37 (56.92) | 25 (45.46) | 0.645 ** (1.000) | 1.95 (0.30–14.50) |
| GG | 3 (4.62) | 4 (7.27) | Reference | |
| AA + GG | 28 (43.08) | 30 (54.55) | Reference | |
| AG | 37 (56.92) | 25 (45.45) | 0.286 * (1.000) | 1.59 (0.77–3.27) |
| AA + AG | 62 (95.38) | 51 (92.72) | Reference | |
| GG | 3 (4.62) | 4 (7.28) | 0.814 ** (1.000) | 0.62 (0.09–3.84) |
| AG + GG | 40 (61.54) | 29 (52.72) | Reference | |
| AA | 25 (38.46) | 26 (47.28) | 0.431 * (1.000) | 0.70 (0.34–1.44) |
| Allele G | 43 (33.10) | 33 (33.00) | Reference | |
| Allele A | 87 (66.90) | 77 (77.00) | 0.710 * (1.000) | 0.87 (0.50–1.50) |
*, Chi-square with Yates correction; **, Fisher’s exact test; %, percentage; 95%CI, 95% confidence interval; n, number of individuals; OR, odds ratio. p-value for Hardy–Weinberg equilibrium: (a) 0.105; (b) 0.127; (c) 0.070; (d) 0.830. Values below 0.05 indicate that the sample is out of Hardy–Weinberg equilibrium. Bold type indicates the presence of statistically significant differences in the proportions of genotypes or alleles between the groups of patients and controls. An alpha error of 0.05 was adopted in the statistical analysis. p-corrected was adjusted using Bonferroni correction for multiple comparisons.
Table 4.
Comparative association of the variant rs3812718 in Sodium Voltage-Gated Channel Alpha Subunit 1 (SCN1A) gene and susceptibility to temporal lobe epilepsy and Online Archive of Brazilian Mutations (ABraOM, from Portuguese Arquivo Brasileiro On-line de Mutações) controls.
| Genotypes and Alleles | Patients n (%) a | Controls n (%) b | p-Value (p-Corrected) | OR (95%CI) |
|---|---|---|---|---|
| GG | 30 (25.00) | 245 (20.90) | 0.144 * (1.000) | 1.54 (0.90–2.62) |
| GA | 60 (50.00) | 549 (46.90) | 0.211 * (1.000) | 1.37 (0.87–2.17) |
| AA | 30 (25.00) | 377 (32.20) | Reference | |
| GG + AA | 60 (50.00) | 622 (53.10) | Reference | |
| GA | 60 (50.00) | 549 (46.90) | 0.579 * (1.000) | 1.13 (0.78–1.65) |
| GG + GA | 90 (75.00) | 794 (67.80) | Reference | |
| AA | 30 (25.00) | 377 (32.20) | 0.131 * (1.000) | 0.70 (0.46–1.08) |
| GA + AA | 90 (75.00) | 926 (79.00) | Reference | |
| GG | 30 (25.00) | 245 (21.00) | 0.357 * (1.000) | 1.26 (0.81–1.95) |
| Allele A | 120 (50.00) | 1303 (50.66) | Reference | |
| Allele G | 120 (50.00) | 1269 (49.34) | 0.898 * (1.000) | 1.03 (0.79–1.34) |
| Genotypes and Alleles | Drug-Resistant n (%) c | Controls n (%) | p-Value (p-Corrected) | OR (95%CI) |
| GG | 17 (26.15) | 245 (20.90) | 0.090 * (1.000) | 2.01 (0.96–4.22) |
| GA | 35 (53.85) | 549 (46.90) | 0.084 * (1.000) | 1.85 (0.97–3.54) |
| AA | 13 (20.00) | 377 (32.20) | Reference | |
| GG + AA | 30 (46.15) | 622 (53.10) | Reference | |
| GA | 35 (53.85) | 549 (46.90) | 0.334 * (1.000) | 1.32 (0.80–2.18) |
| GG + GA | 52 (80.00) | 794 (67.80) | Reference | |
| AA | 13 (20.00) | 377 (32.20) | 0.055 * (1.000) | 0.53 (0.28–0.98) |
| GA + AA | 48 (73.85) | 926 (79.00) | Reference | |
| GG | 17 (26.15) | 245 (21.00) | 0.396 * (1.000) | 1.34 (0.76–2.37) |
| Allele A | 61 (46.90) | 1303 (50.66) | Reference | |
| Allele G | 69 (53.10) | 1269 (49.34) | 0.458 * (1.000) | 1.16 (0.82–1.65) |
| Genotypes and Alleles | Drug-Responsive n (%) d | Controls n (%) | p-Value (p-Corrected) | OR (95%CI) |
| GG | 13 (23.64) | 245 (20.90) | 0.810 * (1.000) | 1.18 (0.56–2.47) |
| GA | 25 (45.45) | 549 (46.90) | 0.897 * (1.000) | 1.01 (0.54–1.90) |
| AA | 17 (30.91) | 377 (32.20) | Reference | |
| GG +AA | 30 (54.55) | 622 (53.10) | Reference | |
| GA | 25 (45.45) | 549 (46.90) | 0.945 * (1.000) | 0.94 (0.55–1.63) |
| GG + GA | 38 (69.09) | 794 (67.80) | Reference | |
| AA | 17 (30.91) | 377 (32.20) | 0.959 * (1.000) | 0.94 (0.53–1.69) |
| GA + AA | 42 (76.36) | 926 (79.00) | Reference | |
| GG | 13 (23.64) | 245 (21.00) | 0.754 * (1.000) | 1.17 (0.62–2.21) |
| Allele A | 59 (53.60) | 1303 (50.66) | Reference | |
| Allele G | 51 (46.40) | 1269 (49.34) | 0.607 * (1.000) | 0.89 (0.60–1.30) |
| Genotypes and Alleles | Drug-Resistant n (%) c | Drug-Responsive n (%) d | p-Value (p-Corrected) | OR (95%CI) |
| GG | 17 (26.15) | 13 (23.64) | 0.439 * (1.000) | 1.71 (0.62–4.76) |
| GA | 35 (53.85) | 25 (45.45) | 0.264 * (1.000) | 1.83 (0.76–4.44) |
| AA | 13 (20.00) | 17 (30.91) | Reference | |
| GG + AA | 30 (46.15) | 30 (54.55) | Reference | |
| GA | 35 (53.85) | 25 (45.45) | 0.464 * (1.000) | 1.40 (0.68–2.89) |
| GG + GA | 52 (80.00) | 38 (69.09) | Reference | |
| AA | 13 (20.00) | 17 (30.91) | 0.245 * (1.000) | 0.56 (0.24–1.29) |
| GA + AA | 48 (73.85) | 42 (76.36) | Reference | |
| GG | 17 (26.15) | 13 (23.64) | 0.916 * (1.000) | 1.14 (0.41–2.63) |
| Allele A | 61 (46.90) | 59 (53.60) | Reference | |
| Allele G | 69 (53.10) | 51 (46.40) | 0.365 * (1.000) | 1.31 (0.79–2.18) |
*, Chi-square with Yates correction; %, percentage; 95%CI, 95% confidence interval; n, number of individuals; OR, odds ratio: (a) 1.000; (b) 0.228; (c) 0.808; (d) 0.816. Values below 0.05 indicate that the sample is out of Hardy–Weinberg equilibrium. An alpha error of 0.05 was adopted in the statistical analysis. p-corrected was adjusted using Bonferroni correction for multiple comparisons.
For rs2910164, the frequencies of the homozygous genotype (CC) (15.00% vs. 9.65%, p-value = 0.043) and C allele (37.80% vs. 29.97%) were superior in patients with TLE compared to controls with a higher risk for the disease [Odds Ratio (OR) = 1.89 (95% confidence interval (95%CI) = 1.06–3.37); OR = 1.38 (95%CI = 1.04–1.82), respectively] (Table 2). Drug-responsive patients also presented higher frequencies of the CC genotype [(21.81% vs. 9.65%; OR = 2.58 (95%CI = 1.25–5.30)] and C allele [(39.09% vs. 29.97%; OR = 1.50 (95%CI = 1.01–2.22)] compared to controls (Table 2).
For rs2298771, the frequency of the heterozygous genotype (AG) (51.67% vs. 40.40%) was superior in patients with TLE compared to controls with a higher risk for the disease [OR = 2.42 (95%CI = 1.08–5.41)] (Table 3). In addition, drug-resistant patients also presented a higher AG genotype frequency [(56.92% vs. 40.40%; OR = 3.36 (95%CI = 1.04–17.30)] compared to the control group. However, for drug-responsive patients, the genotypes and prevalence of alleles were similar in patients and controls (Table 3). For rs3812718, the genotypes and prevalence of alleles were similar in both studied groups (Table 4).
In addition, no differences occurred in the association between the groups of patients, namely the drug-resistant and drug-responsive groups.
2.3. microRNA (miR)-146a (MIR-146A) and Sodium Voltage-Gated Channel Alpha Subunit 1 (SCN1A) Quantification Considering Single Nucleotide Variants (SNVs) rs2910164 and rs2298771
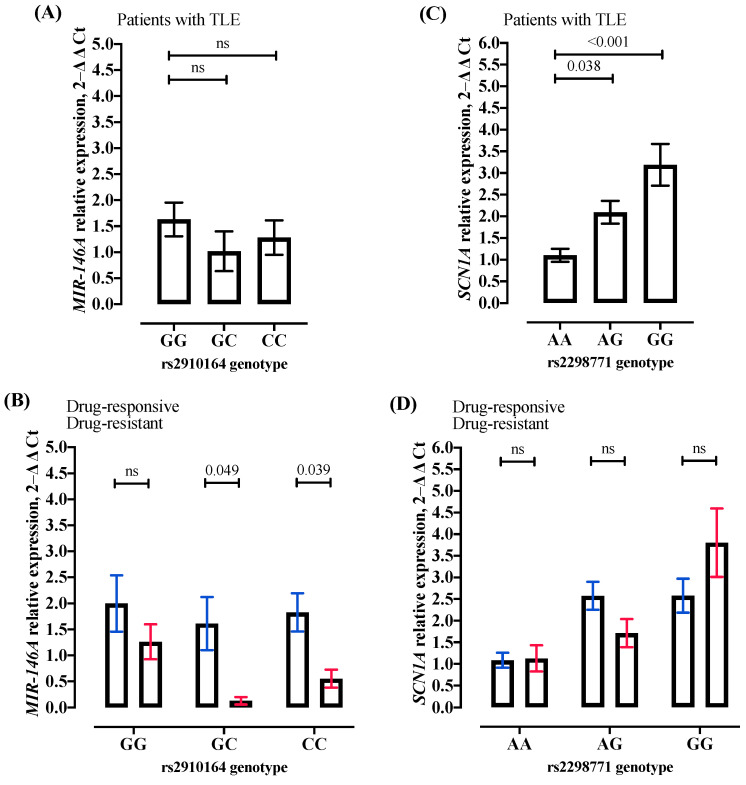
The MIR-146A relative expression levels in samples of TLE patients were lower in GC (1.02 vs. 1.63, p-value = 0.234] and CC (1.03 vs. 1.63, p-value = 0.491) genotypes compared to the GG genotype (Figure 1A); however, no significant association was observed. Moreover, when patients were divided into drug-resistant and drug-responsive groups, the MIR-146A relative expression level was lower in drug-resistant compared to drug-responsive patients for GG (1.3 vs. 2.0, p-value = 0.275), and it was only significantly lower for GC (1.6 vs. 0.1, p-value = 0.049) and CC (1.8 vs. 0.6, p-value = 0.039) genotypes (Figure 1B), possibly indicating an increased NF-kB inflammation process in drug-resistant patients harboring GC or CC genotypes for SNV rs2910164.
Figure 1.
Quantitative real-time polymerase chain reaction (qRT-PCR) for microRNA (miR)-146a (MIR-146A) and Sodium Voltage-Gated Channel Alpha Subunit 1 (SCN1A) gene expression. (A) MIR-146A gene expression from patients with temporal lobe epilepsy (TLE) for each genotype for the single nucleotide variant (SNV) rs2910164 [GG (n = 12); GC (n = 10); CC (n = 7)]. (B) MIR-146A gene expression from patients with drug-responsive and drug-resistant TLE for each genotype for SNV rs2910164 [drug-responsive: GG (n = 6); GC (n = 6); CC (n = 4); drug-resistant: GG (n = 6); GC (n = 4); CC (n = 3]. (C) SCN1A gene expression from patients with TLE for each genotype for SNV rs2298771 [AA (n = 10); AG (n = 9); GG (n = 6)]. (D) SCN1A gene expression from patients with drug-responsive and drug-resistant TLE for each genotype for SNV rs2298771 [drug-responsive: AA (n = 6); AG (n = 4); GG (n = 3); drug-resistant: AA (n = 4); AG (n = 5); GG (n = 3]. All values are represented as mean ± standard deviation. The gene expression was evaluated in patient epileptogenic tissues or peripheral blood from patients with TLE. The statistical analysis was performed using T-Test. Also, all p-values lower than 0.042 were significant after the correction for multiple tests using the Bonferroni approach. 2−∆∆Ct algorithm, the delta–delta cycle threshold; ns, no significant; n, number of individuals (samples).
In contrast, the SCN1A relative expression levels in samples from TLE patients were significantly higher in the AG genotype [2.09 vs. 1.10, p-value = 0.038] and GG (3.19 vs. 1.10, p-value < 0.001) compared to the AA genotype (Figure 1C). The results may indicate an SCN1A loss of function in patients with TLE harboring AA genotypes for SNV rs2298771. In addition, drug-responsive patients presented no significant SCN1A expression levels for AA (1.1 vs. 1.1; p-value = 0.894), AG (2.6 vs. 1.7; p-value = 0.107), and GG (2.6 vs. 3.8, p-value = 0.237) genotypes compared with drug-resistant patients (Figure 1D).
The MIR-146A and SCN1A relative expression values for each evaluated sample are presented in Supplementary Table S1.
3. Discussion
In the present preliminary report, we performed a case–control study to analyze two potentially functional SNVs of the MIR-146A and SCN1A genes and the risk of epilepsy in a Brazilian cohort sample. Previous studies have suggested that decreased miR-146a expression may be associated with increased NF-kB inflammation and susceptibility to the development of epilepsy [11,12]. Moreover, MIR-146A was observed with an increased expression level in human brain astrocytes, and it may inhibit target genes related to epileptic inflammatory process [31].
Only three previous studies have evaluated the SNV rs2910164 and TLE. The first study observed that the rs2910164 variant in the pre-miR146a gene is unlikely to influence the risk of developing TLE or its severity in Italian patients [16]. In the second study, Cui et al. (2015) concluded that the rs2910164 variant was not associated with TLE [18]. The third study from our group suggested that the GC genotype for SNV rs2910164 appears to be associated with susceptibility to drug-resistant TLE in Brazilian patients, probably due to the decreased MIR-146A expression, favoring the NF-kB pathway [11]. The CC genotype for SNV rs2910164 could also be related to susceptibility to drug-resistant TLE, but there was a small number of patients harboring the CC genotype. Thus, to confirm the results, we evaluated a higher number of drug-resistant TLE patients. The same cohort of drug-resistant patients evaluated previously was included in the present study. Additionally, more than 15 drug-resistant patients were also added to the present study. In Boschiero et al. (2020), only three drug-resistant patients were identified as harboring the CC genotype for SNV rs2910164 [11]; here, only three drug-resistant patients were identified as the CC genotype and no risk for TLE was observed, indicating that a larger Brazilian cohort should be evaluated for SNV rs2910164. The hospital at Bragança Paulista (São Paulo, Brazil) is not known for its high complexity, and it is not prepared to receive many difficult-to-manage patients. Moreover, the Brazilian population is mixed, with their origins mainly being from Europeans, Amerindians, Africans, Levantines, and East Asians, explaining our contrasting results [32,33]. Thus, the discrepancy in our results might be due to the ethnic variation and differences in the number of recruited patients.
Corroborating with our previous publication [11], the MIR-146A expression level was lower in drug-resistant compared to drug-responsive patients for GC and CC genotypes, indicating an increased NF-kB inflammation process in drug-resistant patients harboring the GC or CC genotypes. Interestingly, when drug-responsive TLE patients were evaluated, the CC genotype was related to the disease susceptibility compared to control individuals with a 2.6-fold risk for drug-responsive patients with TLE. In fact, the MIR-146A expression level was lower in drug-responsive patients harboring the CC variant, indicating increased NF-kB.
The α subunit of the voltage-gated sodium channel is a large protein of 2000 amino acids and its function is to generate a brief influx of sodium ions by transiently opening in response to neuronal membrane depolarization and closing within milliseconds [34]. Single amino acid substitutions can alter numerous components of channel function and deviate from normal channel function, causing clinical consequences such as epilepsy [35]. Thus, our study evaluated the SNVs rs2298771 and rs3812718, two of the most common SNPs in the intron and exon of the SCN1A gene, influencing the regulation of the gene expression and its structure and functionality, respectively; also, both SNVs are closely related to resistance to sodium-channel-blocking antiepileptic drugs [19].
Two studies have found a correlation between the A allele and combined genotypes GA + AA for SNV rs2298771 and the poor response to antiepileptic sodium channel blockers [19,22]. In contrast, five studies have found no association between genotypes from SNV rs2298771 and sodium channel blocker metabolism or resistance [20,21,23,24,25].
To our knowledge, our study is the first to evaluate the SNV rs2298771 in TLE patients. Interestingly, we found that both the AA genotype and A allele present an increased risk for the disease in drug-responsive patients. In fact, the SCN1A expression level was lower in drug-responsive patients harboring the AA genotype. Our results seem to indicate that the wild-type genotype for SNV rs2298771 in SCN1A, which encodes the voltage-gated sodium channel—NaV1.1 sodium channel alpha subunit—results in a loss-of-function protein presenting a lower expression, with an insufficiency of NaV1.1, as observed in the SCN1A gene mutated in Dravet Syndrome, as well as milder phenotypes associated with genetic epilepsy with febrile seizures plus [34,36,37].
Seven studies have evaluated the susceptibility for general epilepsy seizure, multidrug resistance, and the SNV rs3812718 [19,20,22,23,26,27,28]. Only one study has analyzed TLE risk and the SNV rs3812718 [23]. Thus, the authors compared genotypes and allele frequencies between South Indian Ancestry patients with mesial TLE with hippocampal sclerosis (mTLE-HS) and observed that the AA genotype and A allele were overrepresented in these patients, contributing to increased susceptibility to mTLE-HS [23]. Only one study has demonstrated an association between the genotype harboring the A allele for the SNV rs3812718 and the need to administer higher doses of anti-epileptic drugs than those with the GG genotype, whereas a correlation with the multidrug resistance phenotype was not detectable [26]. Our present study demonstrated no susceptibility to TLE, drug-responsive, or drug-resistant patients, for the SNV rs3812718.
Generally, the cohort of patients with seizure onset during childhood exhibit a worse response to medication. This underscores the need for a deeper understanding of epilepsy as a phenotype reflecting developmental-aging processes characterized by maladaptive neuroplasticity. These processes begin before conception and continue through pregnancy, childhood, and subsequent critical periods such as adolescence and reproductive senescence, posing risks across the lifespan [38].
Over the past decade, significant progress has been made in understanding the genetic and morphogenic mechanisms underlying cortical malformations and developmental brain tumors. Focal malformations are primarily caused by somatic variants in genes associated with cell growth, particularly in the mechanistic Target of Rapamycin (mTOR) pathway for focal cortical dysplasia type 2, acquired in neuronal progenitors during neurodevelopment. Conversely, developmental brain tumors arise from somatic variants in genes linked to cell proliferation, such as the Mitogen-activated protein (MAP)-kinase pathway in ganglioglioma, affecting proliferating glioneuronal precursors. The timing and specific gene involved during neurodevelopment determine the nature and size of the lesion, whether it manifests as a developmental malformation or a brain tumor [39].
SNPs are genetic variations that can influence susceptibility to diseases, including epilepsy. These SNPs may impact gene regulation, protein function, and other biological processes relevant to the development and pathogenesis of epilepsy. Therefore, SNPs could potentially play a role in determining individual susceptibility to epilepsy, including drug-resistant forms, and may contribute to the complex genetic interactions underlying this condition.
Thus, in our drug-responsive cohort, the CC variant genotype of SNP rs2910164 in the MIR-146A gene is more prevalent and patients exhibit lower MIR-146A expression, particularly in genotypes GC and CC, suggesting increased NF-kB-mediated inflammation in drug-responsive cases harboring the CC genotype. For SNP rs2298771 in the SCN1A gene, drug-resistant patients more frequently have the AG genotype versus AA + GG. Higher expression levels of SCN1A are observed in AG and GG genotypes, indicating that the wild-type genotype may lead to a loss-of-function NaV1.1 protein in drug-resistant cases harboring AG or GG genotypes. Our results need to be confirmed in a larger drug-responsive and drug-resistant cohort. In conclusion, the rs2910164-CC and/or rs2298771-AG genotypes are exerting significant risk influence, respectively, on responsive disease and resistant disease, probably due to an upregulated NF-kB and SCN1A loss of function.
4. Materials and Methods
4.1. Research Ethics Committee
This study was approved by the Ethics Committee of São Francisco University (approval no 45723615.0.0000.5514).
4.2. Temporal Lobe Epilepsy Patient Selection and Control Population
The selection of patients with epilepsy was performed from an electronic medical record system at the hospital. Thus, a total of 70 patients with TLE were enrolled, with 55 being drug-responsive and 15 being drug-resistant. For the TLE diagnosis and seizure classification, we evaluated the personal and family history of epilepsy, clinical and neurological physical examination, electroencephalogram and/or video-electroencephalogram, magnetic resonance imaging, or computed tomography. In addition, this study comprised 50 samples of human drug-resistant TLE tissues obtained from surgical amygdalohippocampectomy patients between January 2015 and December 2018. All epilepsy paraffin-embedded tissues were donated from Prof. Dr. Luciano de Souza Queiroz, Department of Pathology, University of Campinas, São Paulo, Brazil. The patients’ inclusion criteria were based on the International League Against Epilepsy [30,40,41,42].
The control population for the association study comprised 1171 healthy blood donor individuals from the ABraOM database, a repository containing genomic variants of the Brazilian population, with a total of 77,236,632 variants (September 2020) [43].
4.3. Deoxyribonucleic Acid (DNA) Samples and Single Nucleotide Variants (SNVs) Identification
Ten milliliters of peripheral venous blood was collected from 55 drug-responsive and 15 drug-resistant patients. Further, genomic deoxyribonucleic acid (DNA) samples for genotyping were isolated using lithium chloride extraction [44]. Genomic DNAs were previously isolated from 50 drug-resistant patient tissues [11] using a phenol- and chloroform-based protocol [45].
The MIR-146A (rs2910164) and SCN1A (rs2298771 and rs3812718) genotypes were identified using real-time polymerase chain reaction (RT-PCR) performed on the StepOne RT-PCR (Applied Biosystems®, Waltham, MA, USA) using the standard TaqMan® genotyping assay (rs2910164: C_15946974_10; rs2298771: C_11748767_20; rs3812718: C_25982233_10) according to the manufacturer’s instructions.
4.4. Quantitative Real-Time Polymerase Chain Reaction
Total ribonucleic acid (RNA) was isolated using Trizol® Reagent (Invitrogen™, Carlsbad, CA, USA) from peripheral blood or paraffin-embedded epileptic tissue samples [rs2910164: GG n = 12; GC n = 10; CC n = 7; rs2298771: TT n = 10; CT n = 9; CC n = 6] according to the manufacturer’s instructions. MIR-146A (assay 000468) and U6 (assay 001973) complementary DNA (cDNA) were synthesized from total RNA according to the TaqMan® real-time assays protocol (Applied Biosystems®). The relative expression of each target was quantified by the delta–delta cycle threshold (ΔΔCt) method [46]. Each sample was examined in triplicate and the raw data are presented as the relative quantity of the target, normalized by U6. For SCN1A analyses, cDNA conversion from total RNA was performed using a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems®, USA). Each sample was examined in triplicate and the expression of each gene was normalized by the control gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and calculated by applying the 2−ΔΔCt method. The MIR-146A and SCN1A expression means for each genotype for the SNVs rs2910164 and rs2298771 (wild-type; heterozygous; variant) were evaluated by T-Test. Primer sequences used for amplification by quantitative real-time polymerase chain reaction (qRT-PCR) with the SYBRGreen dye (Applied Biosystems®, USA) are as follows: SCN1A (forward) 5′-AGGCTGGAATATCTTTGACGG-3′ and (reverse) 5′-GCCAACTTGAAAACTCGCAG-3′; GAPDH (forward) 5′-CCACTTGATTTTGGAGGGAT-3′ and (reverse) 5′-GCACCGTCAAGGCTGAGAAC-3′.
4.5. Statistical Analyses
The Hardy–Weinberg equilibrium was tested using the Chi-square test. Differences between groups were analyzed using the Chi-square test or Fisher Exact test for categorical data (genotype and allele frequencies). In addition, the comparison between groups for numerical data was performed using the T-test (gene expression) and Mann–Whitney test (patients’ age and age of onset). The normality of the numerical data was evaluated using the Shapiro–Wilk test and the Kolmogorov–Smirnov test. For all statistical tests, significance is two-sided and achieved when p-values are less than 0.05. Moreover, a correction for multiple comparisons was performed using the Bonferroni approach. The corrected p-values are presented only in the tables and figures legends. All tests were performed using the Statistical Package for the Social Sciences (IBM Corp. Released 2012. IBM SPSS Statistics for Windows, Version 21.0. Armonk, NY, USA: IBM Corp.). The figure was drawn using the GraphPad Prism version 10.0.0 for Mac, GraphPad Software, Boston, MA, USA, www.graphpad.com (accessed on 16 March 2024).
5. Conclusions
For SNV rs2910164 (MIR-146A), a significantly increased frequency of variant genotype (CC) was observed in drug-responsive patients with TLE. The MIR-146A relative expression level was lower in drug-resistant compared to drug-responsive patients for GC and CC genotypes versus the GG genotype for SNV rs2910164, indicating an increased NF-kB inflammation process in drug-resistant patients harboring GC or CC genotypes.
For SNV rs2298771 in the SCN1A gene, an increased frequency of the AG genotype versus AA + GG genotypes was observed in drug-resistant patients with TLE. The SCN1A relative expression level was higher in AG and GG genotypes versus the AA genotype for SNV rs2298771, indicating that the wild-type genotype of SCN1A, which encodes the NaV1.1 sodium channel alpha subunit, might result in the SCN1A loss-of-function protein presenting a lower expression with an NaV1.1 insufficiency.
Acknowledgments
We thank all the patients and healthy donors for their participation in this study.
Abbreviations
Confidence interval (CI); deoxyribonucleic acid (DNA); glyceraldehyde-3-phosphate dehydrogenase (GAPDH); mesial temporal lobe epilepsy (mTLE); mesial temporal lobe epilepsy with hippocampal sclerosis (mTLE-HS); Messenger RNA (mRNA); Micro-Ribonucleic Acid (microRNA); MicroRNA (miR); nuclear factor-kappa B (NF-kB); odds ratio (OR); polymerase chain reaction (PCR); real-time polymerase chain reaction (RT-PCR); quantitative real-time polymerase chain reaction (qRT-PCR); Online Archive of Brazilian Mutations (ABraOM, from Portuguese Arquivo Brasileiro On-line de Mutações); ribonucleic acid (RNA); single nucleotide variants (SNVs); Sodium Voltage-Gated Channel Alpha Subunit 1 (SCN1A); temporal lobe epilepsy (TLE).
Supplementary Materials
The supporting information can be downloaded at https://www.mdpi.com/article/10.3390/ijms25116005/s1.
Author Contributions
Conception and design: M.M.O.; collection of tissue samples from patients who underwent surgery: L.d.S.Q., P.H.P.d.A. and C.T.P.d.O.; collection of blood samples from patients and healthy controls: R.P.B., K.A.G.T., and A.R.S.; acquisition of data: R.P.B., J.S.d.S., and A.R.S.; analyses and interpretation of data: M.M.O., F.A.L.M., R.P.B. and J.S.d.S.; statistical analyses: F.A.L.M.; drafting of the manuscript: M.M.O. and R.P.B.; study supervision: M.M.O. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Ethics Committee of São Francisco University of (protocol code 45723615.0.0000.5514 and approved on 1 June 2024).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
National Council for Scientific and Technological Development (CNPQ, from Portuguese Conselho Nacional de Desenvolvimento Científico e Tecnológico) scholarship for A.R.S (no 100050/2023-9) and Coordination of Superior Level Staff Improvement (CAPES, from Portuguese Coordenação de Aperfeiçoamento de Pessoal de Nível Superior) scholarship for J.S.S. (no 88887464813/2019-00).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Stafstrom C.E., Carmant L. Seizures and epilepsy: An overview for neuroscientists. Cold Spring Harb. Perspect. Med. 2015;5:a022426. doi: 10.1101/cshperspect.a022426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fiest K.M., Sauro K.M., Wiebe S., Patten S.B., Kwon C.S., Dykeman J., Pringsheim T., Lorenzetti D.L., Jette N. Prevalence and incidence of epilepsy: A systematic review and meta-analysis of international studies. Neurology. 2017;88:296–303. doi: 10.1212/WNL.0000000000003509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World Health Organization (WHO) (2022). Improving the Lives of People with Epilepsy: A Technical Brief. [(accessed on 20 March 2023)]. Available online: https://apps.who.int/iris/bitstream/handle/10665/365270/9789240064072-eng.pdf?Sequence=1.
- 4.Ministério da Saúde do Brasil (2022). Epilepsia: Conheça a Doença e os Tratamentos Disponíveis no SUS. [(accessed on 28 April 2023)]; Available online: https://www.gov.br/saude/pt-br/assuntos/noticias/2022/marco/epilepsia-conheca-a-doenca-e-os-tratamentos-disponiveis-no-sus.
- 5.Semah F., Picot M.C., Adam C., Broglin D., Arzimanoglou A., Bazin B., Cavalcanti D., Baulac M. Is the underlying cause of epilepsy a major prognostic factor for recurrence? Neurology. 1998;51:1256–1262. doi: 10.1212/WNL.51.5.1256. [DOI] [PubMed] [Google Scholar]
- 6.Englot D.J., Morgan V.L., Chang C. Impaired vigilance networks in temporal lobe epilepsy: Mechanisms and clinical implications. Epilepsia. 2020;61:189–202. doi: 10.1111/epi.16423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pal D.K., Pong A.W., Chung W.K. Genetic evaluation counseling for epilepsy. Nat. Neurol. 2010;6:445–453. doi: 10.1038/nrneurol.2010.92. [DOI] [PubMed] [Google Scholar]
- 8.Thomas R.H., Berkovic S.F. The hidden genetics of epilepsy-a clinically important new paradigm. Epilepsy Curr. 2014;10:283–292. doi: 10.1038/nrneurol.2014.62. [DOI] [PubMed] [Google Scholar]
- 9.Jimenez-Mateos E.M., Henshall D.C. Epilepsy and microRNA. Neuroscience. 2013;15:218–229. doi: 10.1016/j.neuroscience.2013.02.027. [DOI] [PubMed] [Google Scholar]
- 10.Schulte U., Thumfart J.O., Klocker N., Sailer C.A., Bildl W., Biniossek M., Dehn D., Deller T., Eble S., Abbass K., et al. The epilepsy-linked Lgi1 protein assembles into presynaptic Kv1 channels and inhibits inactivation by Kvbeta1. Neuron. 2006;49:697–706. doi: 10.1016/j.neuron.2006.01.033. [DOI] [PubMed] [Google Scholar]
- 11.Boschiero M.N., Camporeze B., Santos J.S.D., Costa L.B.D., Bonafé G.A., Queiroz L.S., Van Roost D., Marson F.A.L., de Aguiar P.H.P., Ortega M.M. The single nucleotide variant n.60G>C in the microrna-146a associated with susceptibility to drug-resistant epilepsy. Epilepsy Res. 2020;162:106305. doi: 10.1016/j.eplepsyres.2020.106305. [DOI] [PubMed] [Google Scholar]
- 12.Buainain R.P., Boschiero M.N., Camporeze B., de Aguiar P.H.P., Marson F.A.L., Ortega M.M. Single-nucleotide variants in microRNAs sequences or in their target genes might influence the risk of epilepsy: A review. Cell. Mol. Neurobiol. 2022;42:1645–1658. doi: 10.1007/s10571-021-01058-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Macfarlane L.A., Murphy P.R. MicroRNA: Biogenesis, function and role in cancer. Curr. Genom. 2010;11:537–561. doi: 10.2174/138920210793175895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lukiw W.J., Zhao Y., Cui J.G. An NF-kappaB-sensitive microRNA-146a-mediated inflammatory circuit in Alzheimer disease and in stressed human brain cells. J. Biol. Chem. 2008;283:31315–31322. doi: 10.1074/jbc.M805371200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taganov K.D., Boldin M.P., Chang K.J., Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. USA. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Manna I., Labate A., Mumoli L., Pantusa M., Ferlazzo E., Aguglia U., Quattrone A., Gambardella A. Relationship between genetic variant in pre-microrna-146a and genetic predisposition to temporal lobe epilepsy: A case-control study. Gene. 2013;516:181–183. doi: 10.1016/j.gene.2012.09.137. [DOI] [PubMed] [Google Scholar]
- 17.Li Y., Wang J., Jiang C., Zheng G., Lu X., Guo H. Association of the genetic polymorphisms in pre-microRNAs with risk of childhood epilepsy in a Chinese population. Seizure. 2016;40:21–26. doi: 10.1016/j.seizure.2016.04.011. [DOI] [PubMed] [Google Scholar]
- 18.Cui L., Tao H., Wang Y., Liu Z., Xu Z., Zhou H., Cai Y., Yao L., Chen B., Liang W., et al. A functional polymorphism of the microrna-146a gene is associated with susceptibility to drug-resistant epilepsy and seizures frequency. Seizure. 2015;27:60–65. doi: 10.1016/j.seizure.2015.02.032. [DOI] [PubMed] [Google Scholar]
- 19.Bao Y., Liu X., Xiao Z. Association between two SCN1A polymorphisms and resistance to sodium channel blocking AEDs: A meta-analysis. Neurol. Sci. 2018;39:1065–1072. doi: 10.1007/s10072-018-3308-3. [DOI] [PubMed] [Google Scholar]
- 20.Shi L., Zhu M., Li H., Wen Z., Chen X., Luo J., Lin C., Zhang Z. SCN1A and SCN2A polymorphisms are associated with response to valproic acid in Chinese epilepsy patients. Eur. J. Clin. Pharmacol. 2019;75:655–663. doi: 10.1007/s00228-019-02633-0. [DOI] [PubMed] [Google Scholar]
- 21.Liu M., Mao J., Xu H., Wang J., Zhao P., Xu Q., Du Z. Effects of SCN1A and SCN2A polymorphisms on responsiveness to valproic acid monotherapy in epileptic children. Epilepsy Res. 2020;168:106485. doi: 10.1016/j.eplepsyres.2020.106485. [DOI] [PubMed] [Google Scholar]
- 22.Zhang X., Liu J., Ye J. Association between SCN1A polymorphism and carbamazepine responsiveness in epilepsy: A meta-analysis. Epilepsy Res. 2021;176:106627. doi: 10.1016/j.eplepsyres.2021.106627. [DOI] [PubMed] [Google Scholar]
- 23.Zhao G.X., Zhang Z., Cai W.K., Shen M.L., Wang P., He G.H. Associations between CYP3A4, CYP3A5 and SCN1A polymorphisms and carbamazepine metabolism in epilepsy: A meta-analysis. Epilepsy Res. 2021;173:106615. doi: 10.1016/j.eplepsyres.2021.106615. [DOI] [PubMed] [Google Scholar]
- 24.Mousavi S.F., Hasanpour K., Nazarzadeh M., Adli A., Bazghandi M.S., Asadi A., Rad A., Gholami O. ABCG2, SCN1A and CYP3A5 genes polymorphism and drug-resistant epilepsy in children: A case-control study. Seizure. 2022;97:58–62. doi: 10.1016/j.seizure.2022.03.009. [DOI] [PubMed] [Google Scholar]
- 25.Ashfaq A., Saleem T., Sheikh N., Maqbool H. Genetic analysis of sodium channel genes in pediatric epilepsy patients of Pakistan. Genet. Res. 2022;9:6. doi: 10.1155/2022/1168703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Angelopoulou C., Veletza S., Heliopoulos I., Vadikolias K., Tripsianis G., Stathi C., Piperidou C. Association of SCN1A gene polymorphism with antiepileptic drug responsiveness in the population of Thrace, Greece. Arch. Med. Sci. 2017;13:138–147. doi: 10.5114/aoms.2016.59737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Z.J., Chen J., Chen H.L., Zhang L.Y., Xu D., Jiang W.T. Association between SCN1A polymorphism rs3812718 and valproic acid resistance in epilepsy children: A case-control study and meta-analysis. Biosci. Rep. 2018;38:BSR20181654. doi: 10.1042/BSR20181654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Markovic I., Pejanovic-Skobic N., Bozina N., Susak Sporis I., Sporis D., Basic S. The lack of influence of IVS5-91 G>A polymorphism of the SCN1A gene on efficacy of lamotrigine in patients with focal epilepsy. Neurol. Res. 2019;41:930–935. doi: 10.1080/01616412.2019.1635321. [DOI] [PubMed] [Google Scholar]
- 29.Balan S., Vellichirammal N.N., Banerjee M., Radhakrishnan K. Failure to find association between febrile seizures and SCN1A rs3812718 polymorphism in south Indian patients with mesial temporal lobe epilepsy and hippocampal sclerosis. Epilepsy Res. 2012;101:288–292. doi: 10.1016/j.eplepsyres.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 30.Kwan P., Arzimanoglou A., Berg A.T., Brodie M.J., Allen Hauser W., Mathern G., Moshé S.L., Perucca E., Wiebe S., French J. Definition of drug-resistant epilepsy: Consensus proposal by the ad hoc task force of the ILAE commission on therapeutic strategies. Epilepsia. 2010;51:1069–1077. doi: 10.1111/j.1528-1167.2009.02397.x. [DOI] [PubMed] [Google Scholar]
- 31.Aronica E., Fluiter K., Iyer A., Zurolo E., Vreijling J., Van Vliet E.A., Baayen J.C., Gorter J.A. Expression pattern of mir-146a, an inflammation-associated microRNA, in experimental and human temporal lobe epilepsy. Eur. J. Neurosci. 2010;31:1100–1107. doi: 10.1111/j.1460-9568.2010.07122.x. [DOI] [PubMed] [Google Scholar]
- 32.Pena S.D., Bastos-Rodrigues L., Pimenta J.R., Bydlowski S.P. DNA tests probe the genomic ancestry of Brazilians. Braz. J. Med. Biol. Res. 2009;42:870–876. doi: 10.1590/S0100-879X2009005000026. [DOI] [PubMed] [Google Scholar]
- 33.dos Santos M., Stur E., Maia L.L., Agostini L.P., Peterle G.T., Mendes S.O., Tajara E.H., de Carvalho M.B., Louro I.D., Silva-Conforti A.M. Genetic variability of inflammatory genes in the Brazilian population. Genet. Test. Mol. Biomark. 2013;17:844–848. doi: 10.1089/gtmb.2013.0264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meisler M.H., Hill S.F., Yu W. Sodium channelopathies in neurodevelopmental disorders. Nat. Rev. Neurosci. 2021;22:152–166. doi: 10.1038/s41583-020-00418-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lindy A.S., Stosser M.B., Butler E., Downtain-Pickersgill C., Shanmugham A., Retterer K., Brandt T., Richard G., McKnight D.A. Diagnostic outcomes for genetic testing of 70 genes in 8565 patients with epilepsy and neurodevelopmental disorders. Epilepsia. 2018;59:1062–1071. doi: 10.1111/epi.14074. [DOI] [PubMed] [Google Scholar]
- 36.Meng H., Xu H.Q., Yu L., Lin G.W., He N., Su T., Shi Y.-W., Li B., Wang J., Liu X.-R., et al. The SCN1A mutation database: Updating information and analysis of the relationships among genotype, functional alteration, and phenotype. Hum. Mutat. 2015;36:573–580. doi: 10.1002/humu.22782. [DOI] [PubMed] [Google Scholar]
- 37.Brunklaus A., Brünger T., Feng T., Fons C., Lehikoinen A., Panagiotakaki E., Vintan M.-A., Symonds J., Andrew J., Arzimanoglou A., et al. The gain of function SCN1A disorder spectrum: Novel epilepsy phenotypes and therapeutic implications. Brain. 2022;145:3816–3831. doi: 10.1093/brain/awac210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scher M.S. Interdisciplinary fetal-neonatal neurology training applies neural exposome perspectives to neurology principles and practice. Front. Neurol. 2024;14:1321674. doi: 10.3389/fneur.2023.1321674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blumcke I., Budday S., Poduri A., Lal D., Kobow K., Baulac S. Neocortical development and epilepsy: Insights from focal cortical dysplasia and brain tumours. Lancet Neurol. 2021;20:943–955. doi: 10.1016/S1474-4422(21)00265-9. [DOI] [PubMed] [Google Scholar]
- 40.Fisher R.S., Acevedo C., Arzimanoglou A., Bogacz A., Cross J.H., Elger C.E., Engel J., Jr., Forsgren L., French J.A., Glynn M., et al. A practical clinical definition of epilepsy. Epilepsia. 2014;55:475–482. doi: 10.1111/epi.12550. [DOI] [PubMed] [Google Scholar]
- 41.Fisher R.S., Cross J.H., French J.A., Higurashi N., Hirsch E., Jansen F.E., Lagae L., Moshé S.L., Peltola J., Roulet Perez E., et al. Operational classification of seizure types by the International League Against Epilepsy: Position paper of the ILAE commission for classification and terminology. Epilepsia. 2017;58:522–530. doi: 10.1111/epi.13670. [DOI] [PubMed] [Google Scholar]
- 42.Scheffer I.E., Berkovic S., Capovilla G., Connolly M.B., French J., Guilhoto L., Hirsch E., Jain S., Mathern G.W., Moshé S.L., et al. ILAE classification of the epilepsies: Position paper of the ILAE commission for classification and terminology. Epilepsia. 2017;58:512–521. doi: 10.1111/epi.13709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Naslavsky M.S., Scliar M.O., Yamamoto G.L., Wang J.Y.T., Zverinova S., Karp T., Nunes K., Ceroni J.R.M., de Carvalho D.L., da Silva Simões C.E., et al. Whole-genome sequencing of 1,171 elderly admixed individuals from Brazil. Nat. Commun. 2022;13:1004. doi: 10.1038/s41467-022-28648-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Woodhead J.L., Fallon R., Figueredo H., Langdale J., Malcom A.D.B. Alternative methodology of gene diagnosis. In: Davies K.E., editor. Human Genetic Diseases: A Practical Approach. IRL Press Limited; Oxford, UK: 1986. pp. 51–64. [Google Scholar]
- 45.van Beers E., Joosse S., Ligtenberg M., Fles R., Hogervorst F.B.L., Verhoef S., Nederlof P.M. A multiplex PCR predictor for aCGH success of FFPE samples. Br. J. Cancer. 2006;94:333–337. doi: 10.1038/sj.bjc.6602889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.