Abstract
Adeno-associated virus type 2 (AAV) is known to inhibit the promoter activities of several oncogenes and viral genes, including the human papillomavirus type 16 (HPV-16) E6 and E7 transforming genes. However, the target elements of AAV on the long control region (LCR) upstream of E6 and E7 oncogenes are elusive. A chloramphenicol acetyltransferase assay was performed to study the effect of AAV on the transcription activity of the HPV-16 LCR in SiHa (HPV-positive) and C-33A (HPV-negative) cells. The results reveal that (i) AAV inhibited HPV-16 LCR activity in a dose-dependent manner, (ii) AAV-mediated inhibition did not require the HPV gene products, and (iii) the AAV replication gene product Rep78 was involved in the inhibition. Deletion mutation analyses of the HPV-16 LCR showed that regulatory elements outside the core promoter region of the LCR may not be direct targets of AAV-mediated inhibition. Further study with the electrophoretic mobility shift assay demonstrated that Rep78 interfered with the binding of TATA-binding protein (TBP) to the TATA box of the p97 core promoter more significantly than it disrupted the preformed TBP-TATA complex. These data thus suggest that Rep78 may inhibit transcription initiation of the HPV-16 LCR by disrupting the interaction between TBP and the TATA box of the p97 core promoter.
Adeno-associated virus type 2 (AAV), a member of the Parvoviridae family, has a linear single-stranded DNA genome of 4.7 kb. The AAV genome contains two open reading frames, corresponding to the replication (rep) gene and capsid (cap) gene (5). The larger rep gene products, Rep68 and Rep78, are nearly identical multiple-function proteins. They are involved in AAV DNA replication (31, 33); bind to the specific DNA motif (8, 27, 37); possess endonuclease (21, 22, 34), helicase (21, 35), and ATPase (39) activities; and are pleotropic regulatory proteins of the AAV p5 and p19 promoters (4, 23). Moreover, Rep78 inhibits promoter activities of proto-oncogenes, such as ras (12), c-myb (36), c-fos, and c-myc (14), as well as some viral genes, such as the human immunodeficiency virus (1), human papillomavirus type 16 (HPV-16) (13), and HPV-18 (19) genes. AAV infection is not associated with human diseases (6) but inhibits the oncogenicity induced by its helper viruses, such as adenovirus (11, 29), herpes simplex virus (9), and HPV-16 (15).
A negative correlation between AAV seropositivity and genital cancer has been reported (26), and human cervical cancer is known to be highly associated with HPV-16 and -18 (41). The transformation property of HPV is known to be mediated by its E6 and E7 transforming proteins, which inactivate the tumor suppressor functions of p53 and Rb, respectively (28, 38). The expression of the E6 and E7 transforming proteins is regulated by the long control region (LCR) upstream of the genes encoding these proteins (20). AAV has been found to inhibit the LCR promoter activities of HPV-16 and -18 (13, 19). HPV has thus been implicated as a target of AAV in its suppression of human cervical carcinoma. Hörer et al. (19) reported that several cis elements on the LCR of HPV-18 may be involved in AAV-mediated inhibition of LCR promoter activity. However, the target elements on the HPV-16 LCR required for AAV-mediated inhibition have not been identified.
In this study, we used chloramphenicol acetyltransferase (CAT) assays to investigate AAV-mediated inhibition of HPV-16 LCR promoter activity. The questions of whether HPV gene products are required for the AAV's inhibition, which part of the AAV genome is responsible for its suppression, and where on the HPV-16 LCR promoter the AAV-targeted element is were addressed. We also detected the ability of Rep78 of AAV to disrupt the association of the TATA-binding protein (TBP) with the TATA box of the HPV-16 p97 core promoter.
AAV inhibits the activity of the HPV-16 LCR.
A reporter plasmid (pBL-16LCR-CAT6) used in the later cotransfection assays was first prepared by cloning the wild-type (WT) HPV-16 LCR (from nucleotide 7152 to 103) into the vector pBL-CAT6 (7) to generate pBL-16LCR-CAT6, which contains a cat gene under the control of the HPV-16 LCR. To examine the effect of AAV on HPV-16 LCR promoter activity, this reporter plasmid and an effector plasmid (pAV1) (24) expressing AAV proteins were cotransfected into SiHa human cervical carcinoma cells. When cotransfection experiments were performed, plasmid DNA prepared in a molar ratio, instead of a weight ratio, was used to avoid unequal number of DNA molecules due to the different molecular masses of individual plasmids. For determining the transfection efficiency, a commercially available internal control plasmid was generally used. However, AAV inhibits a wide spectrum of heterologous promoters (16), including the promoters of the commercial plasmids. Alternatively, in the present study, titration experiments using the molar ratios between the effector and reporter plasmids were conducted. In each molar ratio, a pair of samples was prepared. Individual reporter DNA was cotransfected with either the effector pAV1 or the vector pBR322. The promoter CAT activity obtained in the presence of pAV1 was divided by that of pBR322 and is presented as relative CAT activity.
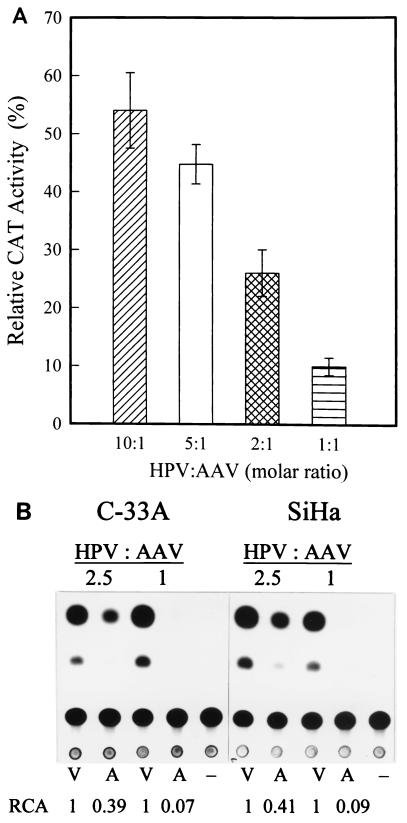
To detect AAV-mediated inhibition of HPV-16 LCR promoter activity, the molar ratios (of HPV-16 LCR to AAV or pBR322) used in cotransfection were 10:1, 5:1, 2:1, and 1:1. The results in Fig. 1A show that the relative CAT activities in extracts of cotransfected cells was reduced from 54.0 to 10.0% when the molar ratio was reduced from 10:1 to 1:1. The results show that the inhibitory effect of AAV was dependent on the relative numbers of HPV-16 and AAV genome copies. The lower the ratio of the HPV-16 LCR to AAV, the higher the AAV inhibitory activity.
FIG. 1.
(A) Inhibitory effect of AAV on HPV-16 LCR promoter activity at different molar ratios. SiHa cells were cotransfected with pBL-16LCR-CAT6 reporter plasmid (10 μg) and either an AAV plasmid (pAV1, which carries WT AAV DNA within the pBR322 vector) (2, 4, 10, or 20 μg) or a control plasmid (pBR322; 1, 2, 5, or 10 μg) to yield HPV/AAV molar ratios of 10:1, 5:1, 2:1, and 1:1. Transfected cells were cultured for 48 h and harvested, and the cell extracts were prepared for CAT assay as described previously (25). Acetylated and nonacetylated spots were quantified by electronic autography with an instant imager (Packard, Meriden, Conn.). The promoter activity in extracts of cells cotransfected with pBL-16LCR-CAT6 and pBR322 was set as 100%. Calculation of the relative activity is described in text. The data are the means ± standard errors of four individual experiments. (B) Representative thin-layer chromatography analysis of the effect of AAV on the HPV-16 LCR in SiHa (HPV-positive) and C-33A (HPV-free) cells. The cells were cotransfected with reporter pBL-16LCR-CAT6 and either effector plasmid pAV1 or vector pBR3224 at molar ratios of 2.5:1 and 1:1. The subsequent steps were the same as described above. The chromatography results presented are from one of three individual experiments. −, pBLCAT6 (negative control); V, pBR322; A, pAV1; RCA, relative CAT activity.
To assess whether HPV gene products are essential for AAV-mediated inhibition, C-33A cervical carcinoma cells without HPV genome integration were cotransfected with pBL-16LCR-CAT and pAV1 for CAT assays. The relative CAT activity in extracts of cotransfected cells decreased from 0.39 to 0.07 when the HPV/AAV molar ratio was adjusted from 2.5:1 to 1:1, respectively (Fig. 1B). The similarity of the results obtained in SiHa and C-33A cells indicates that HPV proteins are not involved in AAV-mediated inhibition.
The AAV replication gene plays a pivotal role in the inhibitory activity of AAV.
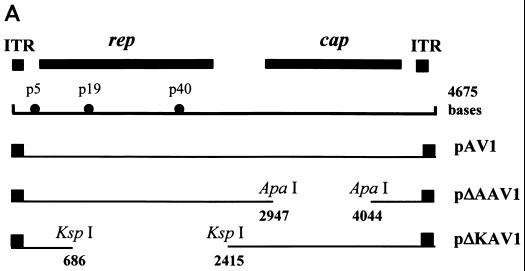
To define which portion of the AAV genome is responsible for the inhibitory activity, individual effector AAV plasmid (WT or mutant) was cotransfected with a reporter plasmid (pBL-16LCR-CAT6) into SiHa cells. Partial-deletion mutants of AAV genomic DNA were obtained by restriction enzyme digestion. Maps of the truncated AAV genomes used are depicted in Fig. 2A. The results of CAT assays demonstrated that WT AAV (pAV1) decreased the HPV-16 LCR activity to 13.2% of its original level (Fig. 2B). A similar result was obtained when the cap gene was deleted (pΔAAV1), indicating that the cap gene is not required for the inhibitory activity of AAV. In contrast, AAV's suppression of the promoter activity of the LCR was reduced to about 50% when the 3′ end of the rep gene was truncated (pΔKAV1). These results suggest that the rep gene plays an important role in AAV's inhibitory function. In addition, our data agree with other findings that Rep78 inhibits the activity of the HPV-16 LCR (14).
FIG. 2.
Inhibitory effects of WT and mutant AAV genomes on activity of the HPV-16 LCR. (A) Plasmid constructs containing the WT or mutated AAV genome. ITR, inverted terminal repeat. (B) SiHa cells were cotransfected with 10 μg of reporter pBL-16LCR-CAT6 and either 5 μg of vector pBR322 or 10 μg of an effector plasmid (pAV1, pΔAAV1, or pΔKAV1) to yield a molar ratio of 2:1. The data are the means ± standard errors of four individual experiments.
The HPV-16 p97 core promoter is inhibited by AAV.
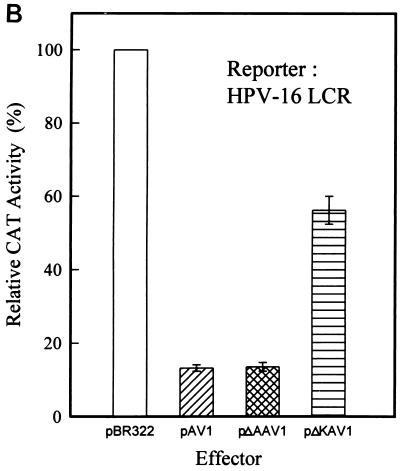
To identify the target elements in the HPV-16 LCR that are involved in the inhibitory activity of AAV, individual constructs containing a WT LCR or LCR fragments (F1 to F7) driving the CAT reporter gene were produced by PCR (Fig. 3A). The sequences of primers used for synthesis of each LCR fragment are listed in Table 1. The transcription activities of the WT and each mutant LCR were initially measured by means of CAT assays (Fig. 3B). The results showed that the promoter activity of the F1 (p97 core promoter) construct was about half that of the WT LCR, while the F2, F3, and F4 constructs showed markedly decreased activity, suggesting the existence of a repressor region between F2 and F4. On the other hand, the promoter activities of F5, F6, and F7 were strikingly increased, 0.5- to ∼4.5-fold, indicating the presence of an enhancer(s) upstream of F4. These results were consistent with a previous report (2) which defined the HPV-16 LCR as the p97 core promoter, silencer, enhancer, and distal regions (Fig. 3A). Furthermore, due to the 4.5-fold-greater activity of F7 than the WT, our results suggest that additional repressors may exist in the distal region.
FIG. 3.
Identification of motifs on the HPV-16 LCR involved in AAV-induced inhibition of promoter activity by mutation analyses. (A) Constructs of the WT and mutant (F1 to F7) HPV-16 LCR. The mutated LCR fragments were produced by PCR as outlined in Table 1 and were subsequently cloned into the vector pBLCAT6. (B) Promoter activities of WT and mutated HPV-16 LCR constructs. SiHa cells were cotransfected with 10 μg of reporter plasmid containing the WT or one of the mutated LCR constructs. The basal CAT activity from cells transfected with the WT construct was defined as 1, and the CAT activity from cells transfected with mutant LCR constructs is reported relative to that of the WT LCR. (C) Inhibitory effects of AAV on the promoter activities of WT and mutated HPV-16 LCR constructs. SiHa cells were cotransfected with 10 μg of a reporter construct carrying the WT or one of the truncated LCR fragments along with pAV1 or pBR322. The HPV/AAV molar ratio used for cotransfection was 2:1. pBLCAT6 and pSV2CAT were used as negative and positive controls, respectively (data not shown). −, not done. The results are reported as percentages of the value relative to the activity of samples from cells transfected with the same LCR construct plus pBR322. The data are the means ± standard errors of three individual experiments.
TABLE 1.
Sequences of the primers used to generate deletion mutant fragments of the HPV-16 LCRa
| End and fragment | Primer | Sequenceb |
|---|---|---|
| 5′ end | ||
| F1 | H16 | CCCAAGCTTGTATAAAACTAAGGGCGTAA |
| F2 | H7855 | CCCAAGCTTAAACCGATTTTGGGTTACAC |
| F3 | H7755 | CCCAAGCTTAAACTTCTAAGGCCAACTAA |
| F4 | H7729 | CCCAAGCTTCCTAATTGCATATTTGGCAT |
| F5 | H7661 | CCCAAGCTTTAAATCACTATGCGCCCACG |
| F6 | H7629 | CCCAAGCTTCTGAATCACTATGTACATTG |
| F7 | H7524 | CCCAAGCTTAACTTGTACGTTTCCTGCTT |
| 3′ end | 100B | AATACGTGGTTTTCTCTTGACCTAGGCGC |
Each primer at the 5′ end was individually coupled with the 3′-end primer to generate LCR fragments F1 to F7 (Fig. 3A). The PCR was performed per the manufacturer's instructions (GeneAmp PCR system 2400; Perkin-Elmer, Branchburg, N.J.). To clone the PCR-amplified fragments into the vector pBLCAT6 at the HindIII and BamHI sites, primers at the 5′ end and 3′ end containing a HindIII and BamHI restriction sites, respectively, were generated. The numbers after H (for HindIII site) in 5′-end primers and before B (for BamHI site) in the 3′-end primer indicate the nucleotide numbers in the HPV-16 sequence.
All 5′-end sequences are in the 5′-to-3′ direction, and the 3′-end sequence is in the 3′-to-5′ direction.
The effects of AAV on the transcription activities of the WT and mutant LCR were subsequently examined by cotransfection of pAV1 effector plasmid and the WT or one of the mutant LCR reporter plasmids into SiHa cells. The results of CAT assays revealed that the relative activity of the WT LCR decreased to 20% in the presence of AAV. Similarly, the activities of F1, F5, F6, and F7 were inhibited by AAV to 17 to 34% (Fig. 3C), indicating that the inhibitory targets of AAV are located not in the enhancer and distal regions of the HPV-16 LCR but within the F1 fragment (p97 core promoter). The inhibitory effects of AAV on F2, F3, and F4 were not scored because the basal transcription activities of these constructs were too low to be meaningfully analyzed in a repression assay.
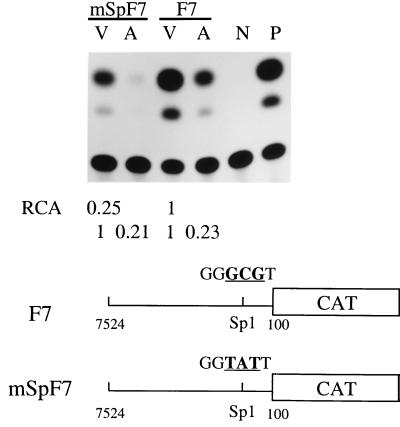
The AAV Rep78 protein has been reported to interact with transcription factor Sp1 and to repress the activity of a synthetic Sp1 binding site-containing promoter (16). To clarify whether the Sp1 binding site within the F1 core promoter region is affected by AAV, the Sp1 binding sequence located at nucleotide 28 to 32 was mutated. The Sp1 binding sequence GGGCGT was changed to GGTATT by PCR and then inserted into the pBL-CAT6 vector as described for Fig. 1 to generate mSpF7. The transcription activity of mSpF7 was reduced to 25% of that of the parental F7 (Fig. 4). This result demonstrates that the Sp1 binding site was mutated successfully and that Sp1 plays an important role in the transcription activity of the p97 core promoter. However, the relative promoter activities of F7 and mSpF7 both decreased to about 20% in cells cotransfected with AAV, suggesting that this Sp1 binding site of the HPV-16 LCR may be not the main target of AAV in the core promoter region.
FIG. 4.
Inhibitory effects of AAV on F7 and mSpF7. SiHa cells were cotransfected with 10 μg of reporter plasmids containing F7 or mSpF7, along with pAV1 (A) or pBR322 vector (V). The molar ratio of HPV to AAV was 2. N and P, negative (pBLCAT6) and positive (pSV2CAT) controls, respectively. The data presented for relative CAT activity (RCA) are the averages of three individual experiments.
Glutathione S-transferase (GST)-Rep78 disrupts the binding of TBP to the HPV-16 p97 core promoter.
We have demonstrated that AAV-mediated repression of HPV-16 LCR promoter activity is related to the p97 core promoter. The p97 promoter contains a TATA box recognized by TBP (32), while AAV Rep78 has been reported to interact with TBP (17). We thus examined whether Rep78 affects the interaction between TBP and the TATA box of the p97 core promoter. Since it is impossible to study the effect of AAV on a TATA sequence-mutated p97 promoter by CAT assay, the interference with the interaction of TBP and the TATA box by Rep78 was alternatively examined by means of electrophoretic mobility shift assays (EMSA).
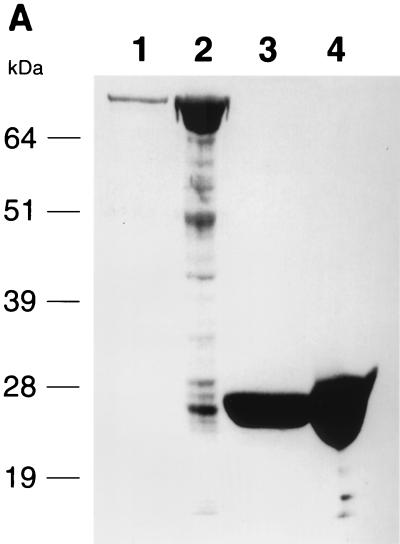
The EMSA were performed under two experimental conditions using recombinant GST-fused Rep78 protein. In assay A, Rep78 protein was added to preformed TBP-16TATA complex; in assay B, Rep78 was incubated with TBP before addition of 16TATA oligonucleotide. The purities of prepared GST and GST-Rep78 proteins are shown in Fig. 5A.
FIG. 5.
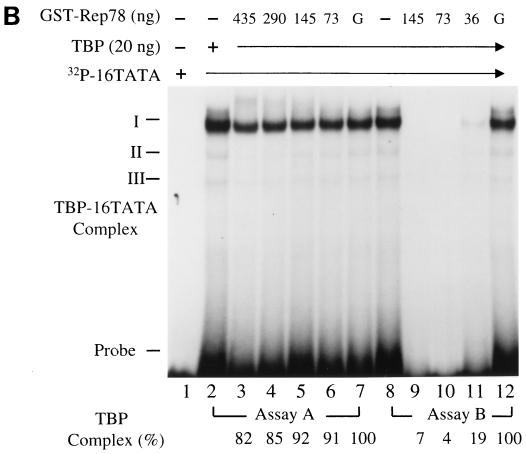
Effect of Rep78 on the complex formation of TBP and the HPV-16 p97 core promoter. (A) Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Coomassie blue staining of GST and GST-Rep78 proteins. The full-length AAV rep gene from nucleotide 321 to 2193 was amplified by PCR and cloned into vector pGEX-5X-3 (Pharmacia Biotech, Uppsala, Sweden) for expression of GST-Rep78 fusion protein. The protocol used for expression and purification of GST-Rep78 was as described by the manufacturer (Pharmacia). Lanes 1 and 3, GST-Rep78 and GST proteins eluted with 10 mM glutathione (reduced form); lanes 2 and 4, GST-Rep78 and GST protein conjugated on glutathione-Sepharose 4B beads. (B) EMSA analysis of the influence of GST-Rep78 on the interaction between TBP and the HPV-16 p97 TATA box. EMSA was performed in a total volume of 20 μl of reaction buffer containing 20 mM HEPES-KOH (pH 7.9), 25 mM KCl, 2 mM MgCl2, 0.1 mM EDTA, 0.5 mM dithiothreitol, 100 μg of bovine serum albumin per ml, 10% glycerol, and 0.025% Nonidet P-40. Twenty nanograms each of human recombinant TBP (Santa Cruz Biotechnology Inc., Santa Cruz, Calif.) and GST-Rep78 at the indicated concentrations was incubated with a 32P-labeled double-stranded 16TATA oligonucleotide (5′-AACGGTTAGTATAAAAGCAGACA), as described by Bauknecht and Shi (3), at 30°C for 30 min. The DNA-protein complexes were resolved on a 5% native polyacrylamide gel in 1× Tris-borate-EDTA (99 mM Tris base, 99 mM boric acid, and 2 mM EDTA [pH 8.3]) running buffer. Lane 1, free 32P-labeled 16TATA oligonucleotide; lanes 2 and 8, TBP-16TATA complexes; lanes 3 to 6, addition of GST-Rep78 to a reaction solution containing preformed TBP-16TATA complexes (assay A); lanes 9 to 11, incubation of TBP with GST-Rep78 prior to addition of 32P-labeled 16TATA oligonucleotide (assay B); lanes 7 and 12, addition of 435 and 145 ng of GST as a negative control for assays A and B, respectively. “G” represents GST and “Probe” represents a free 32P-labeled 16TATA oligonucleotide. The radioactivities of TBP-16TATA complexes on the gel were scored with an instant imager as described for Fig. 1. The percent TBP-16TATA complex formation was calculated as follows: (radioactivity of TBP-16TATA complex in the presence of GST-Rep78/radioactivity of TBP-16TATA complex in the presence of GST) × 100. I, II, and III, TBP-16 TATA complex.
As shown in Fig. 5B, one major and two minor TBP-16TATA complexes were formed in the TBP control (lanes 2 and 8), and these were identified with TBP-specific antibody (data not shown). The addition of GST alone caused about 20% inhibition in TBP-16TATA complex formation (lanes 7 and 12), which was used as the protein control. The TBP-16TATA complex formation was slightly decreased, from 19 to 9% (normalized to GST alone), when GST-Rep78 protein was added (assay A [lanes 3 to 6]). In contrast, GST-Rep78 significantly prevented TBP from binding to 16TATA oligonucleotide; the inhibition increased to 80 to 90% (assay B [lanes 9 to 11]). Obviously, Rep78 is more efficient in inhibiting the binding of DNA to TBP than in abrogating a preformed TBP-DNA complex. Together, these results demonstrated that Rep78 inhibits TBP binding to the HPV-16 p97 core promoter.
The inhibitory effect of Rep78 on the TBP-DNA complex was further confirmed by Western blotting. When TBP was incubated with GST-Rep78, a small amount of degraded TBP was observed while most of TBP remained intact (data not shown). It is thus unlikely that disruption of the TBP-16TATA complex by Rep78 is due to enzymatic degradation mediated by prepared GST-Rep78.
GST-Rep78 inhibits TBP binding to DNA in the presence of Sp1.
The results presented so far demonstrate that Rep78 directly prevented the binding of TBP to the TATA box of the p97 core promoter. A previous report (16) and our unpublished data showed that Rep78 also interacts with Sp1 protein. Our results (Fig. 6, lane 6) showed that within the 16TATA oligonucleotide, Sp1 recognizes an unusual binding site with a GGT sequence (nucleotide 59 to 61) near the TATA box. This agrees with the previously reported finding that Sp1 alternatively interacts with a GGT motif (30). Since Rep78 is known to interact with both TBP and Sp1 (16, 17), the question of whether Rep78 is able to interfere with the binding of TBP to the TATA box of the p97 core promoter in the presence of Sp1 was addressed.
FIG. 6.
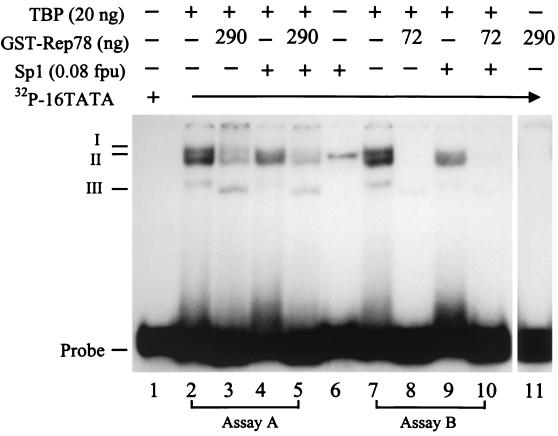
EMSA analysis of the effect of Rep78 on the interaction of TBP and the 16TATA oligonucleotide in the presence of Sp1. EMSA was performed as described for Fig. 5B except that 0.5× TBE running buffer was used. In assay A, GST-Rep78 and Sp1 (Promega Corporation, Madison, Wis.) were added to preformed TBP-16TATA complex individually or mixed (lanes 3 to 5). In assay B, TBP was incubated with GST-Rep78 and Sp1 separately or together prior to the addition of 32P-labeled 16TATA oligonucleotide (lanes 8 to 10). Lane 1, free 32P-labeled 16TATA oligonucleotide; lanes 2 and 7, TBP-16TATA complex; lane 3, addition of GST-Rep78 to the TBP-16TATA complex; lane 4, addition of Sp1 to the TBP-16TATA complex; lane 5, addition of a mixture of GST-Rep78 and Sp1 to the TBP-16TATA complex; lane 6, the Sp1-16TATA complex; lane 8, incubation of TBP with GST-Rep78 prior to the addition of 16TATA oligonucleotide; lane 9, incubation of TBP and Sp1 prior to addition of the 16TATA oligonucleotide; lane 10, incubation of TBP with a mixture of GST-Rep78 and Sp1 before the addition of the 16TATA oligonucleotide; lane 11, incubation of GST-Rep78 with 16TATA probe as a negative control. “Probe” indicates a free 32P-labeled 16TATA oligonucleotide. I, II, and III, TBP-16TATA complex; fpu, footprinting unit.
As shown in Fig. 6, preformed TBP-16TATA complexes were disrupted by GST-Rep78 but affected mildly by Sp1 (lanes 3 and 4, respectively). In the presence of Sp1, GST-Rep78 was still capable of dissociating preformed TBP-16TATA complex (lane 5). Similarly, formation of TBP-16TATA complexes was prevented by GST-Rep78 but partially blocked by Sp1 (lanes 8 and 9, respectively). When GST-Rep78, Sp1, and TBP were mixed prior to addition of 16TATA oligonucleotide, no protein-DNA complex was observed (lane 10). GST-Rep78 alone does not form a complex with 16TATA oligonucleotide (lane 11). These results demonstrated that Rep78 interferes with the binding of TBP to 16TATA oligonucleotide in the presence of Sp1, suggesting that Rep78 interacts with TBP better than with Sp1. Another possibility may be that TBP and Sp1 associate with Rep78 using separate binding sites. The underlying mechanism needs further investigation.
These observations were not due to excess amounts of Rep78 added to the reaction, because the amount of Rep78 used in the assay was titrated to inhibit TBP-16TATA and Sp1-16TATA complex formation (data not shown). Besides, the lower TBP-16TATA signal in the presence of Rep78 may have been due to the degradation of TBP resulting from repeated freeze-thawing or to other, unknown causes.
A similar but not exactly the same TBP-16TATA complex pattern in Fig. 5 and 6 was noted. The reason is not clear, but the similarity is possibly due to repeated freeze-thawing of TBP or to the different ionic strengths of running buffer used. In the assays whose results are shown in Fig. 5 and 6, Tris-borate-EDTA was used at strengths of 1× and 0.5×, respectively. Another unexpected result was obtained in this study. TBP-16TATA complex formation was reduced when Sp1 was added to the reaction. One possible explanation for this observation is the interaction of Sp1 and TBP. It is reported that in the absence of a TATA motif, transcription is initiated through TBP that is anchored to a Sp1-DNA complex formed on the promoter (40).
In summary, AAV-mediated inhibition of transcription activity of the HPV-16 LCR might be due to interference with the binding of TBP to the p97 core promoter by the Rep78 protein, which results in the inhibition of assembly of transcription initiation complexes. Evidence from this study may provide one possible explanation for the ability of AAV to repress a wide spectrum of cellular proto-oncogenes and viral promoters. The transcription activity of a promoter is known to be related to the TATA sequence (TATA box) and its flanking sequences (10, 18). Further studies are needed to determine whether the inhibitory effects of Rep78 are associated with the regulatory composition elements on the target promoter.
Acknowledgments
This work was supported by the National Science Council of the Republic of China (grant NSC87-2314-B-001-015).
We appreciate Ming-Ta Hsu of Yang-Ming University, Taipei, Taiwan, for providing pAV1 and Winston C. Y. Yu of National Health Research Institutes, Taipei, Taiwan, for providing pBL-CAT6 and pBL-16LCR-CAT. We thank Jeff Radcliff of Formosa Medical Editors for reading the manuscript.
Footnotes
This paper is dedicated to the memory of Felicia Wu, who died 19 July 1999.
REFERENCES
- 1.Antoni B A, Rabson A B, Miller I L, Trempe J P, Chejanovsky N, Carter B J. Adeno-associated virus Rep protein inhibits human immunodeficiency virus type 1 production in human cells. J Virol. 1991;65:396–404. doi: 10.1128/jvi.65.1.396-404.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Apt D, Watts R M, Suske G, Bernard H U. High Sp1/Sp3 ratios in epithelial cells during epithelial differentiation and cellular transformation correlate with the activation of the HPV-16 promoter. Virology. 1996;224:281–291. doi: 10.1006/viro.1996.0530. [DOI] [PubMed] [Google Scholar]
- 3.Bauknecht T, Shi Y. Overexpression of C/EBPβ represses human papillomavirus type 18 upstream regulatory region activity in HeLa cells by interfering with the binding of TATA-binding protein. J Virol. 1998;72:2113–2124. doi: 10.1128/jvi.72.3.2113-2124.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beaton A, Palumbo P, Berns K I. Expression from the adeno-associated virus p5 and p19 promoters is negatively regulated in trans by the rep protein. J Virol. 1989;63:4450–4454. doi: 10.1128/jvi.63.10.4450-4454.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berns K I. Parvoviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2173–2197. [Google Scholar]
- 6.Berns K I, Linden R M. The cryptic life of adeno-associated virus. Bioessays. 1995;17:237–245. doi: 10.1002/bies.950170310. [DOI] [PubMed] [Google Scholar]
- 7.Boshart M, Klueppel M, Schmidt A, Schuetz G, Luckow B H R. Reporter constructs with low background activity utilizing the cat gene. Gene. 1992;110:129–130. doi: 10.1016/0378-1119(92)90456-y. [DOI] [PubMed] [Google Scholar]
- 8.Chiorini J A, Weitzman M D, Owens R A, Urcelay E, Safer B, Kotin R M. Biologically active Rep proteins of adeno-associated virus type 2 produced as fusion proteins in Escherichia coli. J Virol. 1994;68:797–804. doi: 10.1128/jvi.68.2.797-804.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cukor G, Blacklow N R, Kibrick S, Swan I C. Effect of adeno-associated virus on cancer expression by herpes virus-transformed hamster cells. J Natl Cancer Inst. 1975;55:957–959. doi: 10.1093/jnci/55.4.957. [DOI] [PubMed] [Google Scholar]
- 10.Das G, Hinkley C S, Herr W. Basal promoter elements as a selective determinant of transcriptional activator function. Nature. 1995;374:657–660. doi: 10.1038/374657a0. [DOI] [PubMed] [Google Scholar]
- 11.de la Maza L M, Carter B J. Inhibition of adenovirus oncogenicity in hamsters by adeno-associated virus DNA. JNCI. 1981;67:1323–1326. [PubMed] [Google Scholar]
- 12.Hermonat P L. Inhibition of H-ras expression by the adeno-associated virus Rep78 transformation suppressor gene product. Cancer Res. 1991;51:3373–3377. [PubMed] [Google Scholar]
- 13.Hermonat P L. Adeno-associated virus inhibits human papillomavirus type 16: a viral interaction implicated in cervical cancer. Cancer Res. 1994;54:2278–2281. [PubMed] [Google Scholar]
- 14.Hermonat P L. Down-regulation of the human c-fos and c-myc proto-oncogene promoters by adeno-associated virus Rep78. Cancer Lett. 1994;81:129–136. doi: 10.1016/0304-3835(94)90193-7. [DOI] [PubMed] [Google Scholar]
- 15.Hermonat P L, Plott R T, Santin A D, Parham G P, Flick J T. Adeno-associated virus Rep78 inhibits oncogenic transformation of primary human keratinocytes by a human papillomavirus type 16-ras chimeric. Gynecol Oncol. 1997;66:487–494. doi: 10.1006/gyno.1997.4789. [DOI] [PubMed] [Google Scholar]
- 16.Hermonat P L, Santin A D, Batchu R B. The adeno-associated virus Rep78 major regulatory/transformation suppressor protein binds cellular Sp1 in vitro and evidence of a biological effect. Cancer Res. 1996;56:5299–5304. [PubMed] [Google Scholar]
- 17.Hermonat P L, Santin A D, Batchu R B, Zhan D. The adeno-associated virus Rep78 major regulatory protein binds the cellular TATA-binding protein in vitro and in vivo. Virology. 1998;245:120–127. doi: 10.1006/viro.1998.9144. [DOI] [PubMed] [Google Scholar]
- 18.Hoopes B C, LeBlanc J F, Hawley D K. Contributions of the TATA box sequence to rate-limiting steps in transcription initiation by RNA polymerase II. J Mol Biol. 1998;277:1015–1031. doi: 10.1006/jmbi.1998.1651. [DOI] [PubMed] [Google Scholar]
- 19.Hörer M, Weger S, Butz K, Hoppe-Seyler F, Geisen C, Kleinschmidt J A. Mutational analysis of adeno-associated virus Rep protein-mediated inhibition of heterologous and homologous promoters. J Virol. 1995;69:5485–5496. doi: 10.1128/jvi.69.9.5485-5496.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Howley P M. Papillomavirinae: the viruses and their application. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2045–2076. [Google Scholar]
- 21.Im D-S, Muzyczka N. The AAV origin binding protein Rep68 is an ATP-dependent site-specific endonuclease with DNA helicase activity. Cell. 1990;61:447–457. doi: 10.1016/0092-8674(90)90526-k. [DOI] [PubMed] [Google Scholar]
- 22.Im D-S, Muzyczka N. Partial purification of adeno-associated virus Rep78, Rep52, and Rep40 and their biochemical characterization. J Virol. 1992;66:1119–1128. doi: 10.1128/jvi.66.2.1119-1128.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Labow M A, Hermonat P L, Berns K I. Positive and negative autoregulation of the adeno-associated virus type 2 genome. J Virol. 1986;60:251–258. doi: 10.1128/jvi.60.1.251-258.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laughlin C A, Tratschin J D, Coon H, Carter B J. Cloning of infectious adeno-associated virus genomes in bacterial plasmids. Gene. 1983;23:65–73. doi: 10.1016/0378-1119(83)90217-2. [DOI] [PubMed] [Google Scholar]
- 25.Lin Y S, Green M R. Similarities between prokaryotic and eukaryotic cyclic AMP-responsive promoter elements. Nature. 1989;340:656–659. doi: 10.1038/340656a0. [DOI] [PubMed] [Google Scholar]
- 26.Mayor H D, Drake S, Stahmann J, Mumford D M. Antibodies to adeno-associated satellite virus and herpes simplex in sera from cancer patients and normal adults. Am J Obstet Gynecol. 1976;126:100–104. doi: 10.1016/0002-9378(76)90472-5. [DOI] [PubMed] [Google Scholar]
- 27.McCarty D M, Pereira D J, Zolotukhin I, Zhou X, Ryan J H, Muzyczka N. Identification of linear DNA sequences that specifically bind the adeno-associated virus Rep protein. J Virol. 1994;68:4988–4997. doi: 10.1128/jvi.68.8.4988-4997.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nevins J R. E2F: a link between the Rb tumor suppressor protein and viral oncoproteins. Science. 1992;258:424–429. doi: 10.1126/science.1411535. [DOI] [PubMed] [Google Scholar]
- 29.Ostrove J M, Duckworth D H, Berns K I. Inhibition of adenovirus-transformed cell oncogenicity by adeno-associated virus. Virology. 1981;113:521–533. doi: 10.1016/0042-6822(81)90180-x. [DOI] [PubMed] [Google Scholar]
- 30.Pereira D J, Muzyczka N. The adeno-associated virus type 2 p40 promoter requires a proximal Sp1 interaction and a p19 CArG-like element to facilitate Rep transactivation. J Virol. 1997;71:4300–4309. doi: 10.1128/jvi.71.6.4300-4309.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Snyder R O, Im D-S, Muzyczka N. Evidence for covalent attachment of the adeno-associated virus (AAV) Rep protein to the ends of the AAV genome. J Virol. 1990;64:6204–6213. doi: 10.1128/jvi.64.12.6204-6213.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tan S H, Gloss B, Bernard H U. During negative regulation of the human papillomavirus-16 E6 promoter, the viral E2 protein can displace Sp1 from a proximal promoter element. Nucleic Acids Res. 1992;20:251–256. doi: 10.1093/nar/20.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tratschin J-D, Miller I L, Carter B J. Genetic analysis of adeno-associated virus: properties of deletion mutants constructed in vitro and evidence for an adeno-associated virus replication function. J Virol. 1984;51:611–619. doi: 10.1128/jvi.51.3.611-619.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walker S L, Wonderling R S, Owens R A. Mutational analysis of the adeno-associated virus Rep68 protein: identification of critical residues necessary for site-specific endonuclease activity. J Virol. 1997;71:2722–2730. doi: 10.1128/jvi.71.4.2722-2730.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Walker S L, Wonderling R S, Owens R A. Mutational analysis of the adeno-associated virus type 2 Rep68 protein helicase motifs. J Virol. 1997;71:6996–7004. doi: 10.1128/jvi.71.9.6996-7004.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walz C, Schlehofer J R. Modification of some biological properties of HeLa cells containing adeno-associated virus DNA integrated into chromosome 17. J Virol. 1992;66:2990–3002. doi: 10.1128/jvi.66.5.2990-3002.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weitzman M D, Kyöstiö S R, Kotin R M, Owens R A. Adeno-associated virus (AAV) Rep proteins mediate complex formation between AAV DNA and its integration site in human DNA. Proc Natl Acad Sci USA. 1994;91:5808–5812. doi: 10.1073/pnas.91.13.5808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Werness B A, Levine A J, Howley P M. Association of human papillomavirus types 16 and 18 E6 proteins with p53. Science. 1990;248:76–79. doi: 10.1126/science.2157286. [DOI] [PubMed] [Google Scholar]
- 39.Wonderling R S, Kyöstiö S R M, Owens R A. A maltose-binding protein/adeno-associated virus Rep68 fusion protein has DNA-RNA helicase and ATPase activities. J Virol. 1995;69:3542–3548. doi: 10.1128/jvi.69.6.3542-3548.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zenzie-Gregory B, Khachi A, Garraway I P, Smale S T. Mechanism of initiator-mediated transcription: evidence for a functional interaction between the TATA-binding protein and DNA in the absence of a specific recognition sequence. Mol Cell Biol. 1993;13:3841–3849. doi: 10.1128/mcb.13.7.3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.zur Hausen H. Papillomavirus infections—a major cause of human cancers. Biochim Biophys Acta. 1996;1288:F55–F78. doi: 10.1016/0304-419x(96)00020-0. [DOI] [PubMed] [Google Scholar]