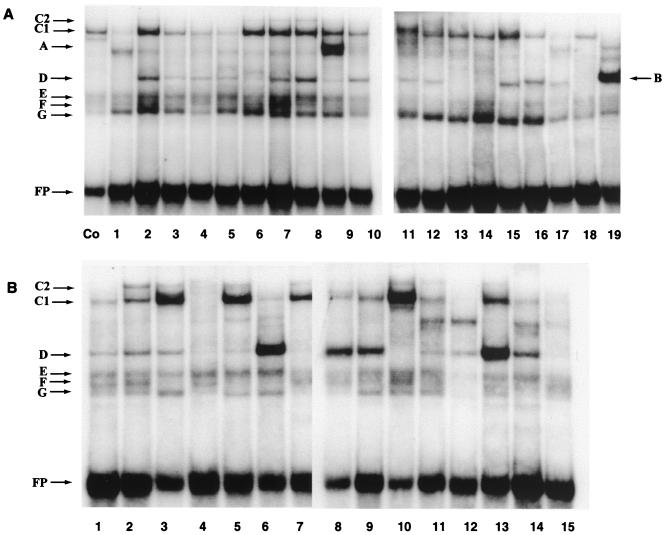
FIG. 5.
EMSA screen of oligonucleotides derived from the 5′-LCR-MAR and the E6-MAR, showing strong binding sites for several nuclear proteins, some of which are common to both MARs. (A) Band shifts obtained with HeLa nuclear extracts and overlapping 31-mer oligonucleotides representing the 5′ third of the HPV-16 LCR from the L1 gene to the E2 binding site most distal from the E6 promoter; (B) band shifts with 40-mer oligonucleotides representing the complete E6 gene. For details, see the footnote to Table 1 and Materials and Methods. The control (Co) is band shift of an oligonucleotide of the CDP/Cut binding site of the gp91phox gene promoter (59). The slowest-mobility complex was termed C1, as it became clear later in this study that it is identical to the complex forming on the PSM, which we termed C1 in a previous study (48). The weak complex C2 that migrated even slower than C1 and is visible only in some slots may be a dimer of the protein giving rise to the C1 complex but could also be a heteromer. It forms efficiently on PSM, which has two flanking binding sites for C1, but only weakly on nonrepeated binding sites. Band B represents binding of YY1 to a previously described site (49), and band A represents binding of the transcription factor USF (data not shown). This study did not make an attempt to identify the proteins giving rise to bands D to G. FP, free probe.