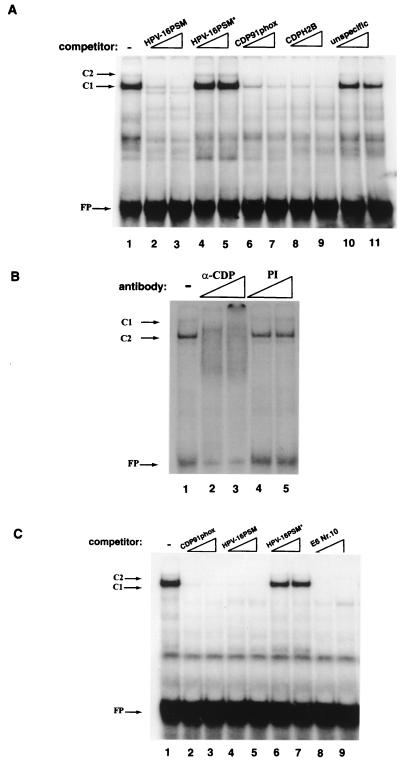
FIG. 7.
Band shift competition and supershift analyses identify CDP/Cut as the factor that binds at least 10 sites in the 5′LCR-MAR and the E6-MAR. (A) E6-10 generates a strong C1 band shift as well as the weak trailing band C2 (lane 1). Formation of both complexes is eliminated by competition with an oligonucleotide representing the HPV-16 PSM (lanes 2 and 3) (47, 48) and the well-studied CDP/Cut binding sites of the promoters of the gp91phox gene (lane 6 and 7) (59) or the histone H2B gene (lanes 8 to 9) (6) but not by a mutant PSM oligonucleotide (lane 4 and 5). As outlined by O'Connor et al. (48), we interpret complex C2 as reflection of a bound dimer binding. FP, free probe. (B) An anti-CDP antibody (α-CDP) abolishes the formation of the C1 and C2 complexes on E6-10. This effect is specific and is not seen upon the addition of a preimmune serum (PI) to the EMSA reaction. (C) LCR-6 produces C1 and C2 complexes similar to those seen with E6-10. The band shift is eliminated by the CDP/Cut binding sites of the gp91phos promoter, the HPV-16 PSM, and E6-10 but not by a mutated PSM oligonucleotide (PSM*). This experiment is representative of numerous crosswise competitions which identify at least five CDP/Cut binding sites in each of the two HPV-16 MARs.