Figure 2.
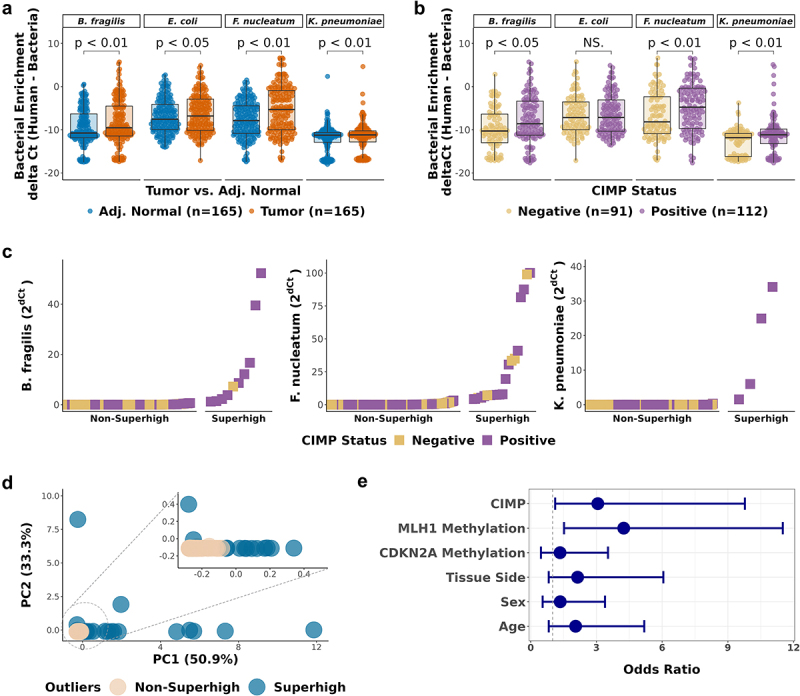
Enrichment of CRC-associated bacteria in CIMP-Positive CRC tumor samples.
a-b. Comparison of bacterial enrichment by qPCR between (a) paired tumor and adjacent normal samples (n = 165), (b) CIMP-Positive and CIMP-Negative tumor samples (n = 203). Significance was calculated by Wilcoxon test. (c) Ranking of the CRC tumor samples by bacterial enrichment. Tumor samples with high bacterial enrichment are classified as Superhigh samples. (d) Principal component analysis (PCA) of the CRC-associated bacteria (B. fragilis, F. nucleatum, and K. pneumoniae) enrichment. The circled area is expanded in the inset PCA plot. (e) Odds ratios of the association of the bacterial Superhigh status, determined by the three bacterial species, with different parameters including the CIMP status, MLH1 methylation, CDKN2A methylation, colorectal tissue side, sex, and age. Blue dots represent the point estimates of the odds ratio, and the lines represent the 95% confidence intervals around the estimates. Fisher’s exact test was used to test for statistical significance for each comparison.
