Figure 4.
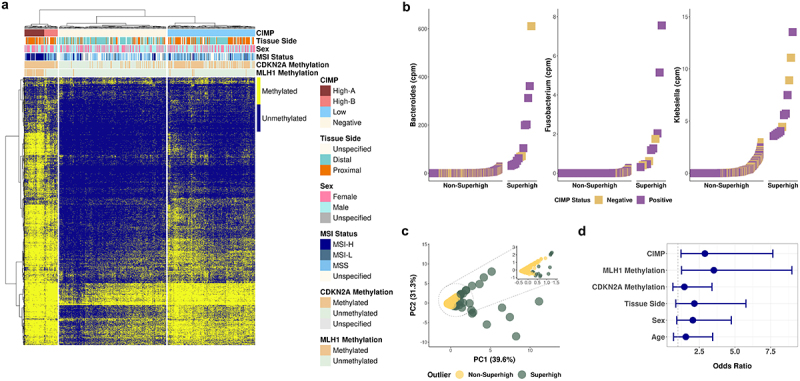
Validation of the CIMP classification method by DREAM using TCGA colon adenocarcinoma and rectum adenocarcinoma methylation array data set.
(a) Unsupervised hierarchical clustering of 393 primary solid tumor and 45 solid tissue normal samples based on 18,289 CpG sites from TCGA COAD and READ 450K methylation array data. (b) Superhigh ranking analysis of bacterial reads from 392 TCGA COAD and READ tumor whole exome sequencing data. (c) PCA of the enrichment of the three CRC-associated bacteria in TCGA datasets. The circled area is expanded in the inset PCA plot. (d) Odds ratios of the association of the bacterial Superhigh cases by TCGA cohorts with different parameters such as the CIMP status, MLH1 methylation, CDKN2A methylation, colorectal tissue side, sex, and age. Fisher’s exact test was used to test for statistical significance for each comparison.
