Figure 5.
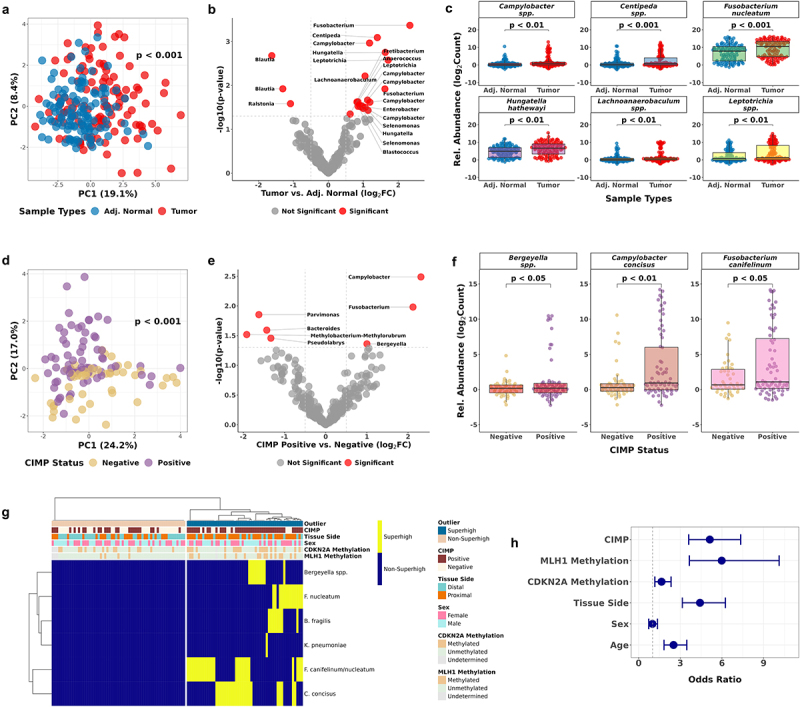
Discovery of CIMP-associated taxa in CRC tumor samples by 16S ribosomal RNA sequencing.
(a) PCA of 23 bacterial taxa selected by the tissue type comparison using ANCOM-BC2. P-values were calculated by PERMANOVA. (b) Volcano plot showing the 23 significant bacterial taxa enriched in tumor or adjacent normal CRC cases. (c) Bacterial enrichment comparison by 16S rRNA gene sequencing paired tumor and adjacent normal samples (n = 228). (d) PCA of 7 bacterial taxa selected by the comparison between CIMP statuses using ANCOM-BC2. (e) Volcano plot showing the 7 significant bacterial taxa enriched in CIMP-Positive or CIMP-Negative tumor samples. (f) Bacterial enrichment comparison between CIMP statuses by 16S rRNA gene sequencing tumor samples (n = 114). (g) Unsupervised hierarchical clustering of the binary Superhigh data of B. fragilis, F. nucleatum, K. pneumoniae, Bergeyella, C. concisus, and F. canifelinum. (h) Odds ratios of the bacterial Superhigh group to be associated with different parameters such as the CIMP status, MLH1 methylation, CDKN2A methylation, colorectal tissue side, sex, and age compared to the Non-Superhigh group. Fisher’s exact test was used to test for statistical significance for each comparison.
