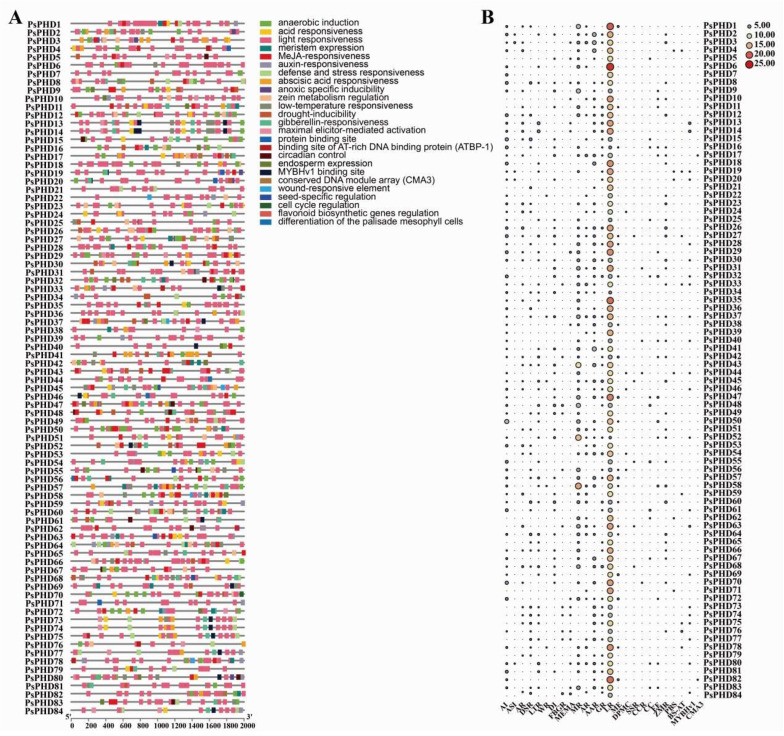
Figure 6.
Cis-acting element analysis of PsPHDs. (A) The 2 kb upstream region of the start codon was analyzed with PlantCARE to identify promoter cis-acting elements, and the results were visualized using TBtools. Diverse promoter cis-acting elements are present in different colored boxes; (B) Heat map of promoter cis-acting element analysis of the PHD finger genes in pea. AI: anaerobic induction; ASI: anoxic-specific inducibility; AR: acid responsiveness; DSR: defense and stress responsiveness; LTR: low-temperature responsiveness; WR: wound-responsive element; DI: drought inducibility; FBGR: flavonoid biosynthetic gene regulation; MEMA: maximal elicitor-mediated activation; MR: MeJA responsiveness; AR: auxin responsiveness; AAR: abscisic acid responsiveness; GR: gibberellin responsiveness; LR: light responsiveness; ME: meristem expression; DPMC: differentiation of the palisade mesophyll cells; SSR: seed-specific regulation; CCR: cell cycle regulation; CC: circadian control; EE: endosperm expression; ZMR: zein metabolism regulation; PBS: protein binding site; BS-AT: binding site of AT-rich DNA binding protein (ATBP-1); MYBHv1: MYBHv1 binding site; CMA3: conserved DNA module array (CMA3).