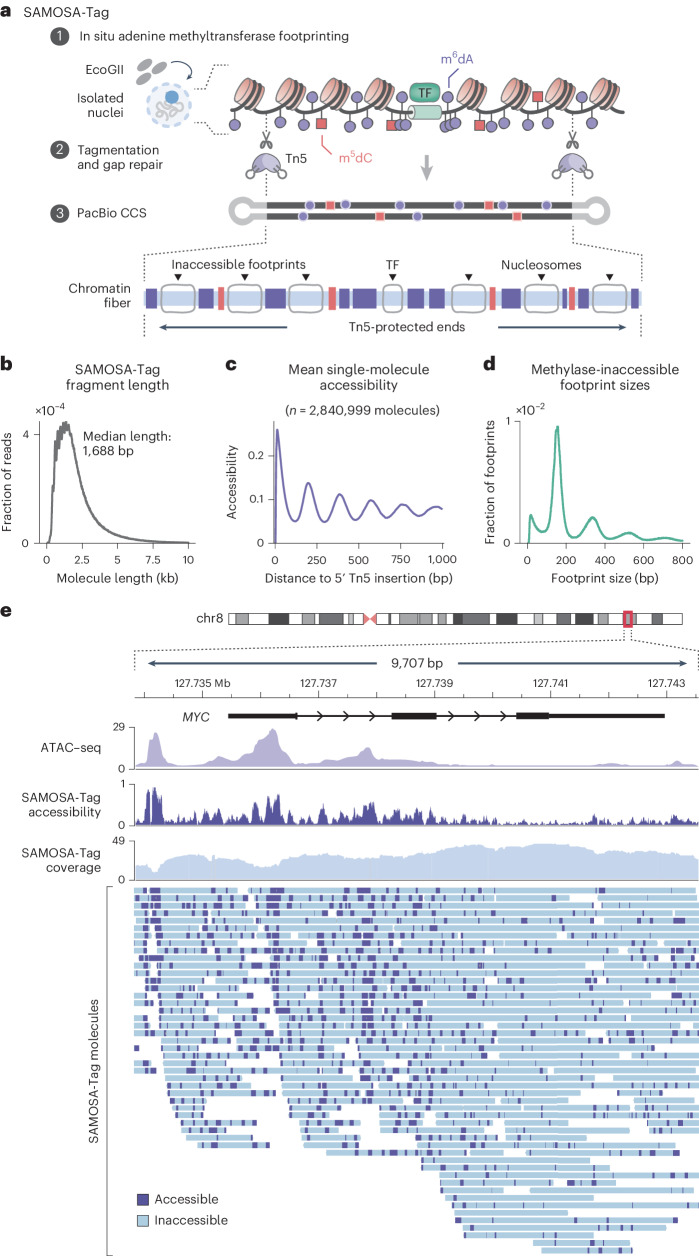
Fig. 3. SAMOSA-Tag: single-molecule chromatin profiling via tagmentation of adenine-methylated nuclei.
a, In SAMOSA-Tag, nuclei were methylated using the non-specific EcoGII m6dAase and tagmented in situ with hairpin-loaded transposomes. DNA was purified, gap-repaired and sequenced, resulting in molecules where the ends resulted from Tn5 transposition, the m6dA marks represented fiber accessibility and computationally defined unmethylated ‘footprints’ captured protein–DNA interactions. b, Length distribution for SAMOSA-Tag molecules from OS152 osteosarcoma cells. c,d, Average methylation from the first 1-kb of molecules (c) and unmethylated footprint size distribution (d) for the same data as in b. e, SAMOSA-Tag fibers at the amplified MYC locus. Predicted accessible and inaccessible bases are marked in purple and blue, respectively. Average SAMOSA accessibility is shown in purple; the matched ATAC–seq track is shown in light purple.