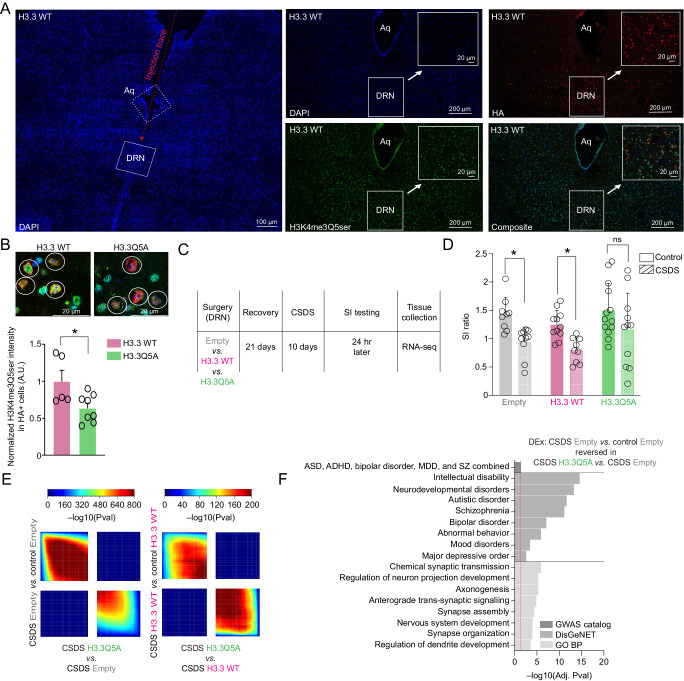
Fig. 4. Viral-mediated downregulation of H3 serotonylation in DRN promotes stress resilience and attenuates stress-induced gene expression.
A IHC/IF images of mouse DRN virally transduced to express HA-tagged H3.3 WT—Far left panel: tiled ×40 image of DRN-containing slice with nuclear co-stain DAPI (4′,6-diamidino-2-phenylindole) showing an example injection trace to target mouse DRN; Middle and far right panels: tiled ×40 images of DRN-containing slice stained for DAPI, HA and H3K4me3Q5ser demonstrating targeted and nuclear expression of H3.3 WT. B Immunofluorescence-based quantification of H3K4me3Q5ser levels/intensity in DRN tissues transduced (HA+) with either H3.3WT (n = 6 slices) or H3.3Q5A (n = 7 slices) (slices analyzed from 3 animals/virus—see Supplementary Fig. 8 for slices used in quantifications). Student’s two-tailed t test revealed a significant difference between H3.3Q5A vs. H3.3 WT transduced mice (p = 0.0444, t11 = 2.269); representative zoomed in ×40 images of quantified DRN cells are provided (co-stained for DAPI, HA and H3K4me3Q5ser). C Experimental timeline for male CSDS after intra-DRN viral transduction by empty vector, H3.3 WT or H3.3Q5A vectors, followed by behavioral testing and tissue collections for RNA-seq. D SI ratios of GFP (n = 9 control; n = 10 CSDS), H3.3 WT (n = 11 control; n = 9 CSDS) and H3.3Q5A (n = 13 control; n = 11 CSDS) transduced mice, control vs. CSDS. Two-way ANOVA significant main effects of stress observed (p = 0.0001, F1,57 = 17.29). Bonferroni’s MC tests within viral group revealed significant differences between control vs. CSDS groups in GFP (p = 0.0310) and H3.3 WT mice (p = 0.0474), with no differences observed between control vs. CSDS H3.3Q5A mice. E Threshold-free RRHO analyses comparing transcriptional profiles for stress-regulated genes in empty vector and H3.3 WT-transduced DRN (control vs. CSDS) to H3.3Q5A-transduced DRN from CSDS mice (n = 4–9/group). Each pixel represents the overlap between differential transcriptomes, with the significance of overlap of a hypergeometric test color-coded. F Pathway enrichment for PCGs displaying differentially expressed genes in CSDS empty vs. control empty comparisons and rescue in CSDS H3.3Q5A vs. CSDS Empty comparisons (FDR < 0.1). Select enriched pathways are shown (FDR < 0.05; Benjamini–Hochberg). For all bar graphs, data presented as mean ± SEM. Source data are provided as a Source Data file.