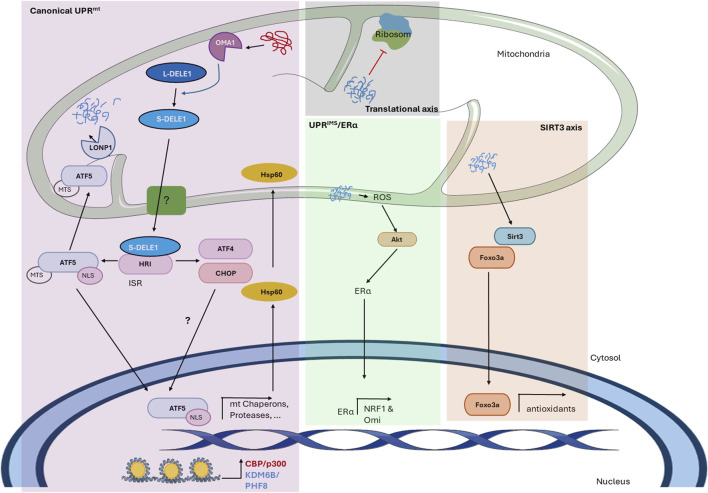
FIGURE 1.
The mitochondrial unfolded protein response in mammals. The figure shows the mechanism of the UPRmt in mammals described thus far. The scheme shows the different axes of the mitochondrial-nuclear retrograde signaling pathway and the proteins involved. In the canonical UPR mt , the accumulation of abnormal proteins within the mitochondrial matrix activates the protease Oma1, which cleaves L-DELE1 into short fragments (S-DELE1) that are released into the cytosol. Once in the cytosol, S-DELE1 interacts with and activates the kinase HRI, activating the ISR and allowing the translation of ATF4, CHOP, and ATF5. The latter is translocated to the nucleus, where it binds to the promoters of UPRmt-related genes following chromatin remodeling by KDM6B and PHF8. CBP/p300 is also involved in epigenetic modification and the expression of UPRmt-related genes. In the translational axis, unfolded proteins reduce the mitochondrial translation rate locally without generating a global response. In the SIRT3 axis, unfolded proteins activate SIRT3, which induces the nuclear localization of the transcription factor FOXO3a to upregulate antioxidant enzyme expression. Finally, the UPR IMS/ ERα is activated by misfolded protein accumulation in the IMS, leading to an increase in ROS levels, which in turn activates Akt kinase to phosphorylate and activate ERα. Undescribed proteins and mechanisms are shown with a question mark (?).