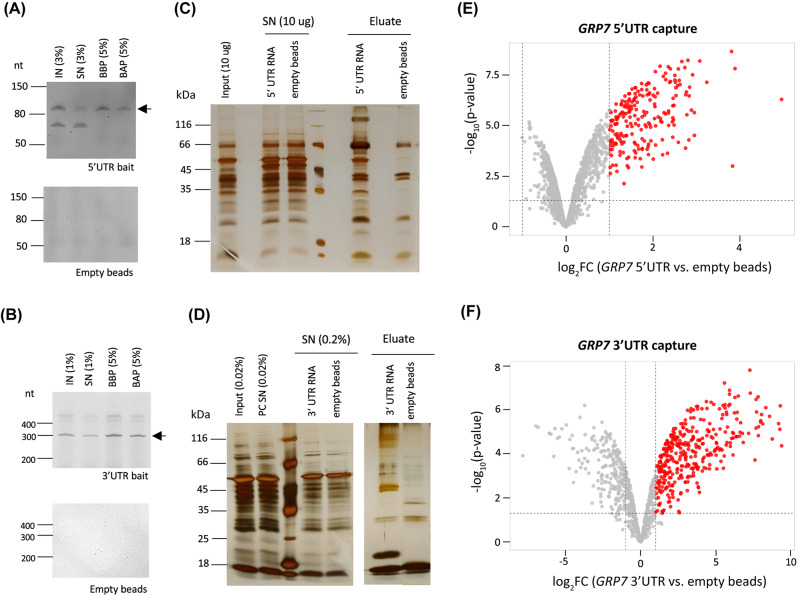
Fig. 6.
Identification of AtGRP7 binding proteins by in vitro pulldowns. (A, B) Coupling efficiency of biotinylated 5’UTR bait (A) and 3’UTR bait (B) to magnetic streptavidin beads. Aliquots of the RNA isolated from the input (IN), supernatant after coupling (SN) and eluates from the beads before (BBP) and after pulldown (BAP) were analyzed on 12.5% urea PAGE gels. Empty beads were used as controls. The arrows indicate the in vitro transcripts of the sizes expected for the 5’UTR (A) or 3’UTR (B), respectively. The additional bands likely represent additional conformations. (C, D) Silver staining of proteins recovered after in vitro captures with 5’UTR bait (C) and 3’UTR bait (D). Aliquots of the input, supernatant and eluates of the respective coupled and empty beads were separated on a 12% SDS-Page and subjected to silver staining. (E, F) Volcano plots of proteins identified by in vitro capture with the 5’UTR bait (E) or 3’UTR bait (F). Significantly enriched proteins (log2 fold change ≥ 1, p-value < 0.05) are indicated by the red dots