SUMMARY
Adaptive immunity provides protection against infectious and malignant diseases. These effects are mediated by lymphocytes that sense and respond with targeted precision to perturbations induced by pathogens and tissue damage. Here, we review key principles underlying adaptive immunity orchestrated by distinct T cell and B cell populations and their extensions to disease therapies. We discuss the intracellular and intercellular processes shaping antigen specificity and recognition in immune activation and lymphocyte functions in mediating effector and memory responses. We also describe how lymphocytes balance protective immunity against autoimmunity and immunopathology, including during immune tolerance, response to chronic antigen stimulation, and adaptation to non-lymphoid tissues in coordinating tissue immunity and homeostasis. Finally, we discuss extracellular signals and cell-intrinsic programs underpinning adaptive immunity and conclude by summarizing key advances in vaccination and engineering adaptive immune responses for therapeutic interventions. A deeper understanding of these principles holds promise for uncovering new means to improve human health.
INTRODUCTION
Adaptive immunity is essential for host protection from infectious and malignant diseases but also contributes to autoimmune and inflammatory disorders under pathophysiological conditions. The adaptive immune system is the collection of cells, factors, and effector mechanisms that, through specialized receptors, recognize and respond to specific antigens, which can be derived from entities outside the body (e.g., pathogens and allergens) or within the body itself (e.g., tumors and self-tissues). The function of the immune system is analogous to the nervous system, with evolved means of sensing, responding to, and remembering the world. The specificity and delayed activation of adaptive immunity contrast with the more rapid and relatively non-specific innate immune response. Further, the adaptive immune system is defined by the emergence of immune memory, the remarkable capacity of lymphocytes to rapidly and precisely respond to a pathogen-derived antigen they encountered before, thereby mediating improved (or complete) protection from re-infection. To accomplish this, the adaptive immune system utilizes an array of diverse “professional” immune cell types that act as individual but interdependent effectors, along with numerous critical interactions with stromal and parenchymal cells throughout tissues.
The core cellular players in adaptive immunity are lymphocytes, specifically T cells and B cells1,2 (Figure 1A). Conventional αβ T cell populations are further classified as CD4+ helper T cells and CD8+ cytotoxic T cells. CD4+ T cells exert multiple effector functions, mediated by both soluble factors and cell-cell interactions. CD8+ T cells act primarily through the killing of specific target cells. B cells secrete soluble effector molecules called antibodies and can also function as antigen-presenting cells (APCs), which present specific antigens to T cells.1 Antibodies bind target antigens with high affinity and interfere with numerous pathogenic processes, such as blocking the attachment of pathogens to host cells and tissue surfaces, among other functions.3 This process prevents pathogen entry into cells, which can provide sterilizing protection against infection from obligate intracellular pathogens. Antibody binding to pathogen surfaces or infected cells also “flags” the cells as targets for innate immune cell-mediated killing, thereby promoting pathogen clearance.
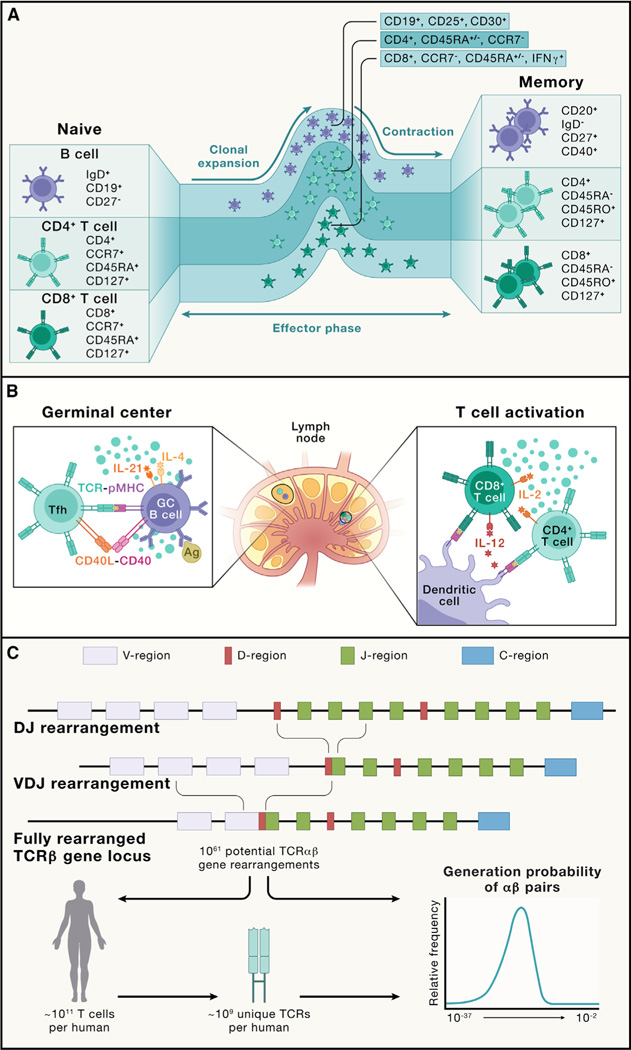
Figure 1. Adaptive immune cell states and the primary response.
(A) B cells, CD4+ T cells, and CD8+ T cells acquire multiple differentiation fates during a normal immune response. Naive T cells and B cells are quiescent cells. Upon activation (not depicted; see B), these cells undergo clonal expansion and acquire effector function (effector phase). Following pathogen control, most effector cells undergo apoptosis (contraction), leaving long-lived memory cells to provide continued immune surveillance (memory). Shown are the major markers used to distinguish various differentiation states or subsets of human T cell and B cell populations discussed in this review.
(B) Primary immune activation occurs in the lymph node. Lymph nodes are spatially organized into discrete B cell and T cell zones. Innate immune cells, especially DCs, process and present antigens to naive CD4+ and CD8+ T cells in the paracortical T cell zone, activating them and inducing clonal expansion. Here, the secretion of IL-12 by DCs drives type 1 differentiation in the responding T cells. IL-2 secretion by CD4+ T cells promotes CD8+ T cell expansion and effector function. Follicular CD4+ T cells also migrate to the B cell zone, where they interact with antigen-specific B cells in the germinal center. Here, B cell recognition of intact antigen via BCRs promotes B cell activation and antigen presentation to CD4+ T cells, which in turn provide activation signals to B cells by CD40L-CD40 interaction and cytokines (e.g., IL-4 and IL-21).
(C) The generation of antigen receptor diversity. TCRs and BCRs are generated by the process of V(D)J recombination, which involves the fusion of separate V, D (on TCRβ and heavy chains), and J segments. Depicted is a TCRβ-chain recombination. Subsequent mRNA splicing brings the constant region together with the V(D)J fusion. This process can result in a vast potential diversity, which is only minimally represented in any individual organism. The distribution of probabilities for an individual TCR to recombine in an individual varies over 20 orders of magnitude.4
Communication between these lymphocytes occurs via two major modalities: secretion of soluble proteins such as cytokines and chemokines and surface ligand-receptor interactions. These signals alter the functional state of the receiving (and sometimes the sending) cell. Collectively, these cell populations, soluble mediators, surface receptors, and their ligands comprise a network of networks, which are overlaid to generate the emergent functions classified as adaptive immunity. Intrinsic redundancy and robustness are key features of the system that maintains function even when deleterious lesions hinder one component. Conversely, numerous feedback loops and inhibitory interactions with immune suppressive populations are also wired in, thereby providing an essential balance that restricts self-reactive and potentially damaging responses to preserve homeostasis and prevent immunopathology.
In this review, we summarize distinct features of adaptive immunity, including how lymphocytes are generated to recognize specific immunological threats (hereafter called threats) and initiate adaptive immunity. We then describe how lymphocytes specialize and adapt to antigen stimulation in both lymphoid and non-lymphoid tissues, including discussing the additional immune stimuli and signaling processes that cooperate with antigen receptor activation to orchestrate adaptive immunity. Importantly, throughout these discussions, the regulatory mechanisms that exist to prevent immunopathology and maintain or re-establish homeostasis are described. Finally, we summarize how adaptive immune responses are harnessed or tailored for therapeutic benefit, including in vaccination against infectious diseases and immune engineering for interventions against cancer and immune-mediated disorders, and close with discussions on emerging and future directions in the field.
HOW DO ADAPTIVE IMMUNE CELLS “SEE” THE WORLD?
B cells and T cells are the essential effectors of adaptive immunity and are exquisitely tuned to target specific pathogens. A cascade of events precedes lymphocyte activation and expansion to ensure that their potent effector function is focused on true pathogenic threats. Here, we discuss the stepwise formation of a primary response and the unique generation of antigen receptors that define the adaptive lymphocyte lineage and mediate antigen recognition. The broad diversity of antigen receptors must be regulated through selection events to limit the development of responses that may damage the host.
Overview of adaptive immune responses
An initial encounter with antigen results in the priming of adaptive immune cells, which subsequently develop into effector cells for immune defense and memory cells to mediate immune memory.5 To understand this process, it is helpful to consider the course of a primary immune response (Figure 1B). For example, after initial infection by a respiratory virus, epithelial cells and local innate immune cells respond over the course of hours to days to attempt control of the pathogen. The lymphocytes that will eventually target this infection are in a quiescent state, circulating through the blood and lymphatics. To initiate this response, dendritic cells (DCs, a type of APCs) and the lymphatic vessels traffic viral antigens to local lymph nodes.6 Here, by secreting chemokines, these DCs will attract naive CD4+ and CD8+ T cells. T cells will then “test” whether they recognize any of the virus-derived antigens “presented” on the DC surface in complex with a host molecule called the major histocompatibility complex (MHC; in humans, these proteins are also called human leukocyte antigens [HLAs]). T cells “see” the peptide-MHC (pMHC) complex via their antigen receptor, called the T cell receptor (TCR), with co-receptors CD4 and CD8 defining T cell subsets recognizing pMHC antigens via different MHC molecules (class II versus class I, respectively).7 Antigen receptors are surface proteins that are the essential sensors of adaptive immune recognition.8,9 Within an organism, each T cell, to a first approximation, contains a unique TCR.10 From any given virus, between 5 and 50 target pMHCs may be recognized, with a corresponding ~250–25,000 naive T cells capable of recognition and becoming activated by binding with an avidity that triggers sufficient TCR signaling.11–13 If a TCR recognizes pMHC on a DC in the appropriate inflammatory conditions, the T cell becomes activated to initiate the subsequent adaptive immune response.
For naive T cells, TCR activation leads to an exit from the quiescent state via the activation of downstream signaling pathways14 and metabolic reprogramming.15 Massive transcriptional, epigenetic, and translational events promote altered surface protein expression and secretion of cytokines and chemokines, which recruit more naive T cells to the local lymph node to permit additional antigenic “screening.” Following quiescence exit, a replication program called clonal expansion is initiated. Consequently, there is a pronounced accumulation of antigen-specific T cells that are normally present at low and variable frequencies, thereby heightening the responses against rapidly dividing pathogens. This replication proceeds via a programmed timer partly regulated by the transcription factor Myc, with the cell undergoing replication as many times as possible within the timer window.16 T cells, once they exit the quiescent state, have among the fastest division times of healthy cells in the mammalian body (estimates of ~10 h for CD4+ and 6–8 h for CD8+ T cells).17–20 Each daughter T cell is similarly activated and carries an identical TCR, thus amplifying the “knowledge” of antigen recognition.
The activation and expansion of a CD4+ T cell response provide support for the activation and expansion of CD8+ T cells in the local lymph node. Co-stimulatory molecules (e.g., CD28 and 4–1BB) and cytokines, such as interleukin (IL)-12 and IL-2 secreted by innate immune cells and CD4+ T cells, enhance CD8+ T cell activation, prolonging division and stimulating specific transcriptional programs. As CD8+ T cells divide and differentiate, they express surface molecules, including ligands for death receptors to induce apoptosis in target infected cells, and also generate inflammatory cytokines and release intracellular stores of cytolytic molecules, including granzymes and perforin.21 After their initial activation and expansion, chemokine gradients guide CD8+ T cells to traffic from the lymph node to the site of infection, where they ultimately execute effector function.22–24
In the same lymph node, naive B cells also test their B cell receptors (BCRs) for binding viral antigens. Rather than pMHC, BCRs recognize whole and intact proteins, and the BCR can bind to three-dimensional conformational epitopes on a viral protein’s surface. Then, BCR-induced signaling, metabolic and transcriptional cascades, comparable to those induced by TCR signaling, activate B cells and initiate differentiation and replication.25 Activated B cells and CD4+ T cells meet at the interface of T cell and B cell zones (T-B border), where BCR-activated B cells can present processed viral proteins as pMHC.26 CD4+ T cells further promote B cell differentiation through additional ligand-receptor interactions (e.g., CD40L-CD40 and ICOS-ICOSL)25 and the secretion of cytokines (e.g., IL-4 and IL-21) that direct class switching to various isotypes (immunoglobulin [Ig]M, IgG, IgA, and IgE).27,28 These interactions promote B cell differentiation into one of two divergent paths. Some B cells can become short-lived antibody-secreting cells called plasmablasts, while others can enter specialized, organized structures called germinal centers.29 In germinal centers, B cells undergo a process called somatic hypermutation, where BCRs are diversified and their specificity altered through the introduction of mutations by the enzyme activation-induced cytidine deaminase (AID).30 Newly mutated BCRs undergo a second round of competitive T-B cell interactions, referred to as selection, in which they must demonstrate their ability to bind and present antigen to specialized germinal center-localized CD4+ T cells.26 Depending on the timing and specific characteristics of the CD4+ T cell and B cell interactions in germinal centers, a second fate divergence occurs in which some “selected” B cells become memory B (Bmem) cells that are capable of responding to subsequent infections, while others become long-lived antibody-producing plasma cells that predominantly reside in the bone marrow.31,32 At full activation, a single plasma cell can produce and secrete thousands of antibodies per second.31
As this primary response matures, coordinated efforts by both localized and systemic lymphocytes and their synergistic effector functions control the pathogen. Following resolution of the infection, >90% of the effector CD4+ and CD8+ T cells, along with a substantial portion of short-lived plasmablast B cells, undergo a regulated process of apoptotic cell death (called contraction). After this contraction phase, there is still a relative increase in the number of antigen-specific B cells and T cells that can recognize the infection that just occurred (as compared with the naive compartment). These cells form the reservoir of memory cells that can be quickly recalled on subsequent challenges, underlying immunity to re-infection.20,32,33
Antigen receptor recognition
The critical recognition event for initiating an adaptive immune response is antigen receptor (TCR or BCR) binding to antigen (pMHC or intact protein epitopes, respectively). How is it possible to seed the naive repertoire (the collection of antigen receptors within an organism) with enough diverse TCRs and BCRs to recognize various epitopes (the antigenic component recognized by the TCR or BCR), including those that are completely novel from an evolutionary perspective? An extremely conservative estimate suggests that at least 109 unique T cell clones exist in the naive repertoire (a number five orders of magnitude larger than the number of genes in the entire genome).10 The solution to this limitation is that each T cell or B cell undergoes a highly coordinated process of somatic recombination to generate a unique antigen receptor with one heavy and one light chain. Mature lymphocytes change their genomes by re-arranging and ligating gene segments to generate novel coding sequences (Figure 1C).34 In humans, TCRs are either αβ or γδ (with the α and γ chains corresponding to the light chains). For BCRs, the heavy chain is referred to as IgH, while the light chain is either Igκ or Igλ. The TCR and BCR loci are extremely large (several hundred kilobases), with regions of multiple gene segment variants. Each chain has both a cassette of “variable,” or V regions, and joining, or “J” regions, with the heavy chain containing an additional cassette of “diversity,” or D regions. To generate a receptor, the DNA is cut by the recombinase activating gene (RAG) enzymes and then stitched back together utilizing a specialized double-stranded break DNA repair process fusing one V, one D (for heavy chains), and one J region.34 DNA repair components Ku, DNA-dependent protein kinase (DNA-PK), Artemis, DNA ligase IV, and XRCC4 all participate in maintaining the proximity of the severed DNA strands and re-ligating them.34 In addition, terminal deoxynucleotidyl transferase (TdT) introduces template-independent nucleotides in the severed junction before ligation and repair, thereby providing an additional source of diversity in receptor generation to recognize a wide range of antigens.34
Collectively, these sources of diversity (gene segment selection, combinatorial diversity, junctional diversity, and the pairing of heavy and light chains) result in an estimated 1061 potentially unique TCRαβ chains.35 This number is exceptionally large—much higher than all of the T cells that have or will ever be generated in humans. Under conditions of random recombination, we would expect that no two T cells would arise within or between individuals with an identical receptor. Although the occurrence of such “public” receptors is uncommon for the full two-chain combination, for single chains, sharing is observed in ~15% of the TCRβ chains.36,37 This observation indicates that the recombination process is strongly biased toward the generation of some receptor chains more than others. A major accomplishment in the last several years, facilitated by the large amount of antigen receptor repertoire data produced by deep sequencing analyses, is the precise calculation of the probability of generating any given TCR or BCR.38–41 These approaches provide reliable estimates of how likely certain receptors are to occur, which can be verified by measuring the degree to which receptors are shared across the population. These tools have multiple applications, including for diagnostics and early detection of diseases, including chronic infections, tumors, and autoimmunity.
Beyond the intrinsically vast diversity available in the naive repertoire, B cells have a further mechanism for diversification of their antigen receptors. In the context of the germinal center structure described earlier, communication between CD4+ T cells and B cells leads to the induction of enzyme AID.30 AID causes the deamination of cytidine, converting it to uracil in the DNA encoding the antigen receptor. The DNA repair pathway then converts this to a thymine or to other bases depending on the repair pathway engaged, thereby introducing a point mutation in the coding sequence. If this mutation enhances the affinity of the BCR for its ligand, the B cell will receive a survival signal. Otherwise, it will undergo apoptosis. This process leads to the affinity maturation of the BCR (and the subsequent secreted antibodies), with the progression of the germinal center reaction resulting in a two-logarithm increase in the measured affinity of secreted antibodies over the course of a response.30
The processes of V(D)J recombination and affinity maturation are intrinsically dangerous for the cell and the host, as they involve the purposeful breakage, re-ligation, and/or mutation of DNA.42 Many pediatric and some adult tumors are driven by fusion genes created by illegitimate V(D)J recombination, demonstrating the embedded potential for malignant transformation in these processes.43 These include chronic myelogenous leukemia, which can contain a translocation between the BCR itself and Abl kinase,44 and T cell acute lymphoblastic leukemia, which can arise from RAG-mediated recombination between two non-antigen receptor genes, SIL and TAL1.45 As a result, antigen receptor recombination is highly regulated by transcriptional induction (both of recombination machinery and at the recombination locus46,47) and the physical sequestration of the antigen receptor loci within the nucleus in “recombination factories.”48
There are many preventive and therapeutic strategies that could arise from harnessing this central means of immune recognition. The ability to decode the repertoire is thus a focus of a number of investigators and is often called “the holy grail” of immunology, though it remains elusive.49 Although we cannot perform a simple mapping between the sequence of an antigen receptor and the identity of its target, significant progress has been made in extracting information from repertoire sequences. Receptors that recognize the same antigens often look very similar to each other.50–52 Finding identical paired chain receptors between individuals (“true public” receptors) is extremely unlikely, but receptors that share a high degree of homology are very common. These conserved motifs strongly suggest that the project of decoding the repertoire is likely to succeed, as it points to a conserved and limited set of solutions underlying how any particular antigen is seen by antigen receptors.53,54 Of note, only 17 epitopes with at least 50 unique paired TCRs are curated in the largest online database to date (accessed and analyzed in January 2024).55 Another complication for the prediction of specificity is the poorly understood extent of cross reactivity possible for any given receptor. That receptors must be cross-reactive is known to be true.56 Estimates as high as one million unique targets per TCR have been made, which imposes further challenges for the decoding problem. However, the translational potential of solving the repertoire code and recent advances in structural modeling keep this problem a major focus of current antigen receptor research.
Balancing self versus non-self recognition: Positive and negative selection
Considering the intrinsically stochastic nature of recombination, the generation of autoreactive TCRs and BCRs seems highly likely. For TCRs, an additional constraint is that they need to recognize peptides in the context of MHC to function properly. How then does T cell development ensure that such conditions (i.e., both lack of autoreactivity and restriction by MHC) are met? For T cells, their development in the thymus includes a rigorous process of both positive and negative selection, often referred to as “thymic education.”57 Following recombination, receptors must bind to MHC to provide a survival signal; if this does not occur, the cell undergoes apoptosis, removing it from the population. A receptor that binds MHC with too strong an affinity may induce apoptosis due to excessive activating signal, which helps eliminate self-reactive specificities from the repertoire (called central tolerance). To allow T cells to properly screen their receptors against antigens that will eventually be seen throughout the body, mechanisms exist to express diverse proteins from every major organ system and tissue in the thymus, with the transcription factor Aire, primarily expressed by medullary thymic epithelial cells, playing a central role in this process.58 Moreover, CD4+ T cells with moderate to high binding affinity for self-antigens are likely to upregulate expression of the transcriptional regulator Foxp3, which endows them the capacity to develop into a distinct lineage known as regulatory T (Treg) cells. After exiting the thymus, Treg cells mediate crucial immunosuppressive function to prevent autoimmunity but also have emerging roles in supporting tissue repair and physiology, and these two effects are described further below. Developing B cells also undergo processes of selection while they are developing in the bone marrow. Self-reactive, immature B cells can be deleted, or alternatively, their BCRs can undergo “receptor editing,” which allows for additional gene recombination events to produce a non-self-reactive BCR.59 Altogether, given the potentially deleterious effects of adaptive immune responses to host tissues, these developmental processes are crucial for “educating” T cells and B cells by equipping them with proper antigen receptors to mediate productive immune responses while avoiding self-disruption.
WHAT KIND OF RESPONSES DOES ADAPTIVE IMMUNITY GENERATE IN THE PERIPHERY?
Appropriate activation of adaptive immunity requires reprogramming of naive lymphocytes into effector cells, which are crucial to defend the host against invading pathogens or to influence other immune or tissue cells (Figure 2). How does the adaptive immune system in the peripheral lymphoid organs (primarily consisting of spleen and lymph nodes) respond to diverse threats initially, re-encounter them after initial clearance, or adapt to constantly facing them? Also, what mechanisms prevent adaptive immunity from carrying out autoimmune reactions in response to self-antigens expressed in the peripheral tissues (called peripheral tolerance)? We discuss effector and memory responses, T cell functional adaptation to chronic antigen stimulation, and Treg-mediated peripheral tolerance and restriction of inflammation (Figure 3), which collectively balance protective immunity against autoimmune reactions.
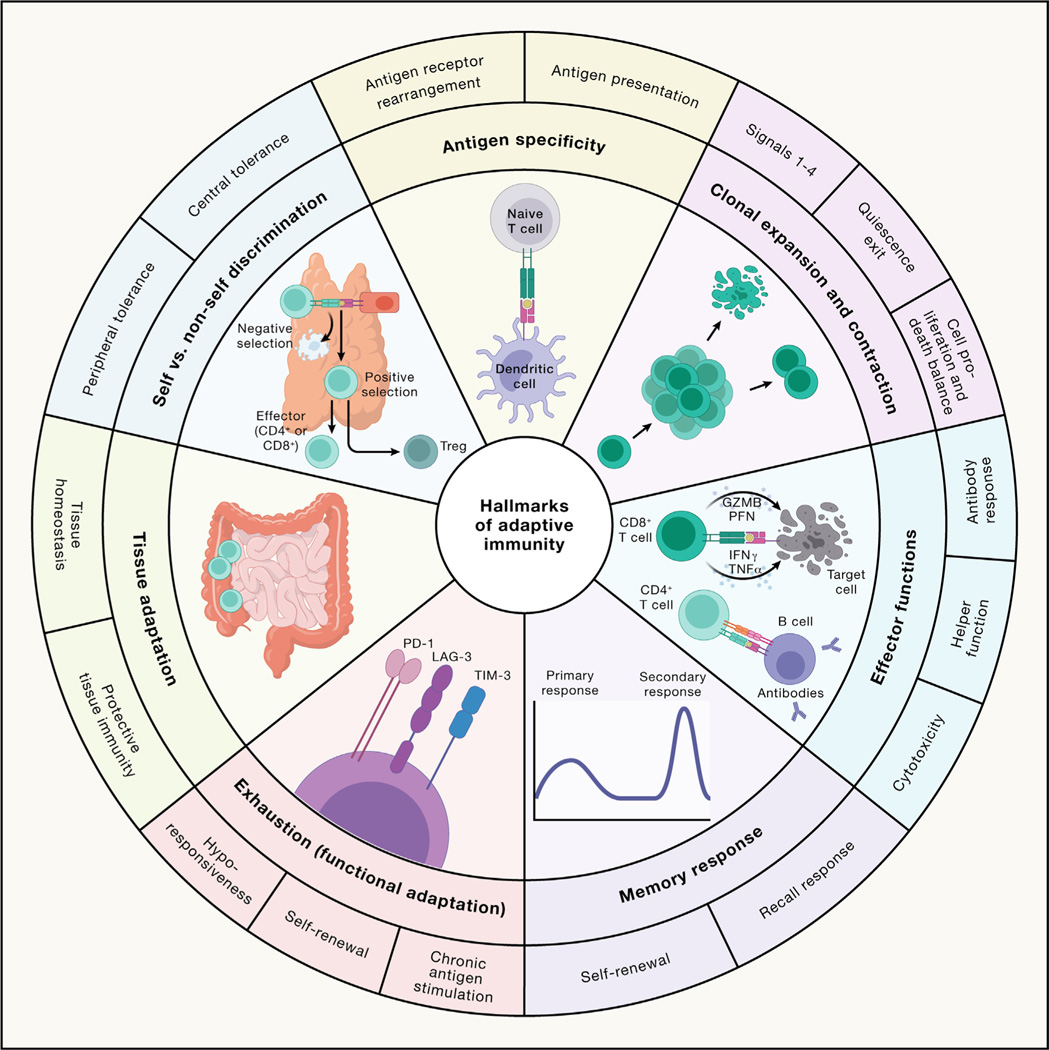
Figure 2. The hallmarks of adaptive immune responses.
The scheme illustrates the seven hallmarks of adaptive immunity described within the article: antigen specificity (contributed by antigen receptor rearrangement and antigen presentation); clonal expansion and contraction (contributed by antigen, co-stimulation, cytokines, and nutrients [signals 1–4, respectively]; quiescence exit; and a balance of cell proliferation and death); effector functions (mediated by B cell antibody responses, CD4+ T cell helper function, and CD8+ T cell cytotoxicity); memory response (contributed by recall response and self-renewal of memory cells); exhaustion or functional adaptation (contributed by chronic antigen stimulation, self-renewal, and hypo-responsiveness); tissue adaptation (presented as protective tissue immunity and tissue homeostasis); and self versus non-self discrimination (contributed by central and peripheral tolerance). PFN, perforin; GZMB, granzyme B.
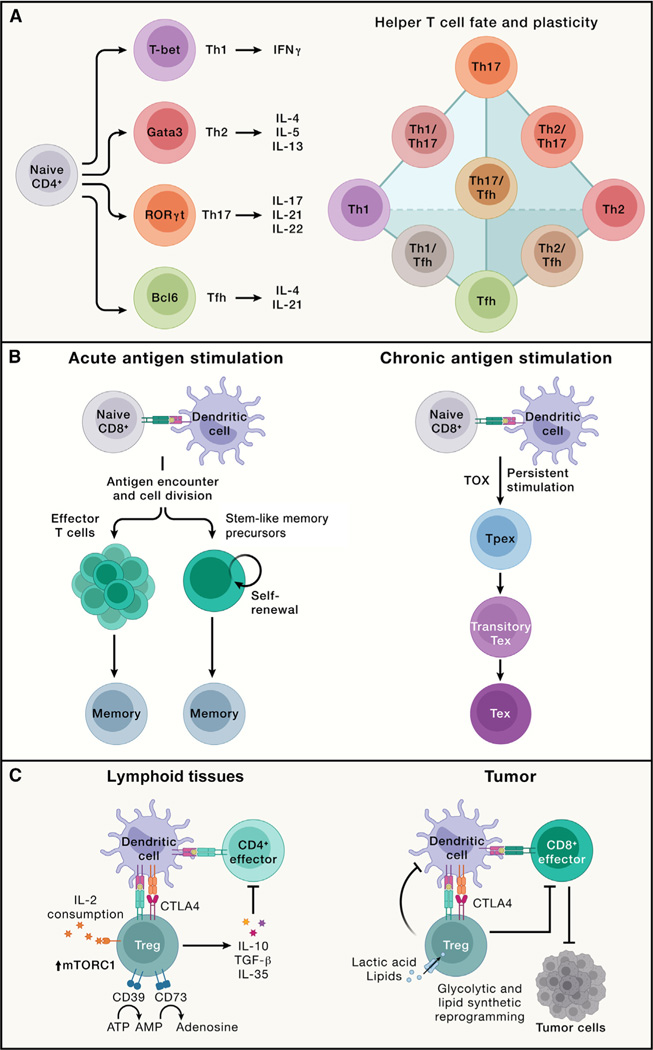
Figure 3. Major types of adaptive immune responses, including effector, memory, exhausted T cell, and regulatory T cell responses.
(A) Left: naive CD4+ T cells become activated via TCR activation combined with co-stimulation and further respond to cytokines in the microenvironment to differentiate into Th1, Th2, Th17, and Tfh cells. These effector T cells express the unique transcription factors T-bet, Gata3, RORγt, and Bcl6, respectively, and produce distinct cytokines. Specifically, Th1 cells produce IFNγ; Th2 cells secrete IL-4, IL-5, and IL-13; Th17 cells synthesize IL-17, IL-21, and IL-22; and Tfh cells generate IL-4 and IL-21. Right: the schematic highlights the representative interconnectedness in the plasticity between CD4+ helper T cell subsets. T helper cells with plasticity can display phenotypes from two distinct CD4+ T cell lineages.
(B) Acute (left) and chronic (right) antigen stimulation results in the development of memory and exhausted CD8+ T cells, respectively. Left: memory CD8+ T cells can de-differentiate from a subset of effector cells or arise directly from stem-like memory precursors that are derived from antigen-stimulated naive T cells. Right: under persistent antigen stimulation, CD8+ T cells upregulate TOX expression and differentiate into Tpex cells, which further become Tex cells. In Tex cells, a transitory Tex population bridges the differentiation between Tpex and Tex cells, although Tex cells may also form without progression through a functional effector state (not depicted).
(C) Left: in lymphoid tissues, thymus-derived Treg cells expressing the transcription factor Foxp3 exert immune suppressive function through multiple mechanisms. Treg cells secrete immunosuppressive cytokines (e.g., IL-10, TGF-β, and IL-35) and express co-inhibitory molecules such as CTLA4 to suppress effector T cells. They also express enzymes CD39 and CD73 that convert pro-inflammatory extracellular ATP to adenosine. The high expression of IL-2 receptors on Treg cells serves as an IL-2 “sink” to dampen IL-2-induced stimulatory effects on NK and CD8+ T cells (not depicted). Both IL-2 signaling and mTORC1 activity are required for Treg suppressive function and metabolic fitness in vivo. Right: Treg cells are a major component of the immunosuppressive tumor microenvironment. Intratumoral Treg cells have unique requirements for nutrients and metabolic programs and may co-localize with a specialized dendritic cell population called cDC1s to suppress the antitumor function of CD8+ T cells.
Diverse effector responses: Responding to different types of threats
How does the immune system respond to the vast diversity of invading pathogens and immunological insults? One solution is the generation of specialized effector cells that display functional diversity based on signals derived from the priming APCs and changing environments. For example, CD4+ T cells respond to pro-inflammatory cues and differentiate into effector cells, including the Th1, Th2, and Th17 subsets (“h” stands for “helper”; namely, the function to help other immune cells such as CD8+ T cells or B cells) of cells that mediate protective immune responses following bacterial, helminth, and fungal infections, respectively. These functions are orchestrated by lineage-specific transcription factors (T-bet, Gata3, and RORγt), signature cytokines, and other effector molecules. In addition, Bcl6-expressing CD4+ T follicular helper (Tfh) cells are crucial for supporting B cell-mediated antibody responses60 (Figure 3A).
However, effector cells are not simply fixed lineages, and they adapt their functions to ever-changing microenvironmental and immunological cues, a phenomenon called plasticity. Of note, Th17 cells display more plasticity than Th1 or Th2 cells. For instance, Th17 cells can adopt IL-23-mediated Th1-like features with enhanced capacity to induce autoimmunity61 and inflammation,62 and mechanistic (or mammalian) target of rapamycin complex 1 (mTORC1)-dependent metabolic rewiring contributes to this process.63 Th17 cells may also transdifferentiate into IL-10-producing cells (Tr1) with immunosuppressive function to resolve inflammation.64 Further, Th17 cells can produce IL-4 in helminth infection65 and possibly allergic asthma.66 Beyond plasticity, effector responses are functionally heterogeneous. For example, homeostatic (non-pathogenic) Th17 cells promote tissue homeostasis and prevention of microbial infection under steady state, especially at mucosal sites such as the intestine, whereas pathogenic Th17 cells precipitate inflammatory and autoimmune diseases.67 Th17 cell pathogenicity is associated with co-expression of IFNγ and granulocyte-macrophage colony-stimulating factor (GM-CSF)67 and requires rewiring of glucose,68,69 lipid,70 and polyamine71,72 metabolism. Moreover, Tfh cell function is tailored in response to specific immunological insults, which supports class switching to various antibody isotypes that provide protective immunity to different types of pathogens.73 The inherent plasticity and heterogeneity of CD4+ T cells for functionally adapting to changing environments may be harnessed for therapeutic interventions.
Activated B cells proliferate and migrate to the T-B border, where they interact with Tfh cells. Tfh cells provide multiple helper signals, including cytokines that direct immunoglobulin class switching (from IgD and IgM to IgG, IgE, or IgA isotypes), resulting in a pool of B cells with diversified functions.25 Recent high-dimensional profiling, especially single-cell RNA sequencing (scRNA-seq), has revealed additional heterogeneity of T cells, B cells, and other adaptive immune cells, including in patient populations with cancer,74,75 rheumatoid arthritis,76 severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection,77 and neurodegenerative diseases.78 For instance, scRNA-seq of patients with bladder cancer79 revealed the existence of clonally expanded cytotoxic CD4+ T cell states. These cells have the capacity for killing autologous tumor cells, and their gene signature predicts response to anti-PD-L1 treatment, further supporting the notion of diversity and heterogeneity of adaptive immune responses.
Immune memory: Remembering the threats upon re-encounter
Immune memory, a hallmark of adaptive immunity (Figure 2), enables a rapid and enhanced immune response upon antigen re-encounter. This effect is associated with an increased frequency of antigen-specific T cells with intrinsically enhanced functionality compared with naive populations and is studied in all lymphocytes, especially CD8+ T cells33 (Figure 3B). Whereas the majority of antigen-specific effector T cells undergo contraction, long-lived CD8+ T cells may descend from a subset of effector cells that retain the capacity to re-acquire stem-like characteristics (i.e., signature molecules expressed by naive T cells).80 This effector to memory cell transition is a “de-differentiation” process orchestrated by epigenetic mechanisms like DNA demethylation.80 Alternatively, memory cells may emerge from a self-renewing population before or during the clonal expansion phase following antigen stimulation,81,82 thereby linking differentiation to replicative history.83 Further, asymmetric partitioning of key signaling molecules during the first cell division may reinforce memory formation.33,84 Transcriptional and metabolic pathways control memory CD8+ T cell differentiation, which require TCF-1,85,86 Myb,87 and Foxo1,88 as well as metabolic processes (e.g., mitochondrial oxidative phosphorylation [OXPHOS] and mitochondrial dynamics89). Future studies using cutting-edge lineage-tracing tools are needed to reconcile or unify these differentiation models and identify the underlying mechanisms.
For an effective recall response to occur upon pathogen rechallenge, memory CD8+ T cells found at the site of infection, in lymphoid organs, and in circulation all participate, with each subset exhibiting differential capabilities for self-renewal, longevity, and cytotoxic function. The memory pool mainly includes central memory (TCM), effector memory (TEM), and tissue-resident memory (TRM) cells, although stem cell-like memory (TSCM) and effector memory re-expressing CD45RA (TEMRA) cells have also been described.33 TCM cells are quiescent memory cells expressing low levels of cytotoxic markers and high levels of lymphoid-tissue homing markers (e.g., CCR7 and CD62L) for their localization to lymphoid tissues. TCM cells may undergo self-renewal in response to IL-7 and IL-15 and differentiate into TEM cells. By contrast, TEM cells display intermediate longevity, express lower levels of these homing markers, and circulate between the blood and inflamed non-lymphoid tissues. Upon re-infection, TEM cells are rapidly recruited from circulation to provide immune defense due to their inherent cytotoxic function.33
Germinal centers support the generation of the memory B cell pool, which forms after an iterative process of BCR diversification and affinity maturation. Two different memory B cell populations emerge: long-lived antibody-secreting plasma cells and Bmem cells.31,32 Long-lived plasma cells produce serum antibodies that can neutralize a pathogen and prevent infection. Bmem cells are rapidly re-activated upon subsequent antigen re-encounter and BCR engagement, causing them to proliferate and rapidly produce expanded populations of antibody-secreting cells.32 Bmem cells circulate and survey the body for their antigen (or a closely related variant), or alternatively take residence in tissues to protect against re-infection. Thus, diversified T cell and B cell subsets are involved in establishing immune memory.
T cell exhaustion: Facing the threats all the time
Although effector lymphocytes function to clear invading pathogens in acute infection, chronic infection, such as HIV, occurs when pathogens are not resolved, leading to persistent antigen stimulation. Under such conditions, T cells gradually become functionally exhausted, a state associated with dampened effector function,90 which is also observed in progressive tumors (Figure 3B). T cell exhaustion prevents tissue damage and excessive immunopathology while also contributing to the curtailment of pathogen and tumor growth, thereby reflecting a hypofunctional state of adaptation.91 Exhausted CD8+ T cells upregulate the expression of co-inhibitory molecules (namely, PD-1, LAG3, TIM-3, and TIGIT); gradually reduce proliferation; and display a unique chromatin landscape compared with effector and memory T cells. TOX is the master transcriptional regulator for inducing the exhaustion program but is largely dispensable for effector or memory T cell formation during acute infection.92–96
Given the functions of CD8+ T cells in mediating adaptive immunity to tumors and chronic infection, the mechanisms underlying T cell exhaustion and how to reinvigorate their functionality are being actively investigated. These efforts led to the discovery that exhausted T cells are separated into two major functional subsets: TCF-1+ precursor exhausted (Tpex) and TCF-1− terminally exhausted T (Tex) cells (Figure 3B). Tpex cells exhibit inherent stemness and self-renewal capacity97–102 and directly respond to antibody-based immunotherapies (called immune checkpoint blockade [ICB]) by producing a proliferative burst of functional CD8+ T cells to control pathogens98–100 or tumors.103,104 Among Tpex cells, a transcriptionally distinct CD62L+ Tpex cell population retains long-term proliferative potential and multipotency in a Myb-dependent manner.105 Transitory exhausted CD8+ T cells (with heightened effector function) are an intermediate state between Tpex and Tex cells.97,106–109 Further, transcriptional regulomes program the stepwise differentiation of exhausted CD8+ T cells, and enforcing Tpex cell quiescence exit and transitory Tex cell accumulation both bolster antitumor effects.109 Of note, antigen-specific T cells may also undergo terminal exhaustion without progression through a functional effector state. The differentiation trajectory of exhausted T cells in discrete chronic infections or tumors warrants further investigation.
Spatiotemporal control of CD8+ T cell responses is evident in tumors. Tumor-draining lymph nodes (tdLNs) contain an abundant Tpex cell population (possibly derived from tumor-specific memory (TTSM) cells110), which requires MHC-I-dependent interaction with type 1 conventional DCs (cDC1s) in the T cell and marginal zones of tdLNs for their maintenance.111–114 When Tpex cells migrate from tdLNs to the tumor, they undergo terminal differentiation. Further, the tertiary lymphoid structure facilitates the influx of B cells and T cells into the tumor site,75 which has been shown to support distinct subsets of exhausted T cells.115,116 Applications of spatial transcriptomics technologies and lineage tracing tools will help decipher cell state transitions and cell-cell interactions underlying lymphocyte adaptation across space and time.
Treg cells limit autoimmunity and inflammation
A defining feature of the adaptive immune system is the ability to distinguish self-tissue antigens from “non-self” (e.g., invading pathogens) (Figure 2). Although central tolerance is crucial for immune homeostasis, it is only partially effective. Therefore, mechanisms of peripheral tolerance exist to prevent autoimmune reactions, including cell-intrinsic mechanisms (e.g., lymphocyte quiescence, ignorance, anergy, and senescence, as reviewed elsewhere117) and extrinsic control, predominantly mediated by Treg cells. Loss of these cells or their lineage-defining transcription factor Foxp3 triggers early-onset fatal autoimmunity in mice and humans,118 suggesting that Treg cells act as a cellular “brake” to prevent adaptive immunity from targeting self-tissue (Figure 3C). Treg cells require Foxp3 for their function and also adapt to enforce context-specific immune regulation mediated by immunosuppressive cytokines (e.g., IL-10, TGF-β, and IL-35118); co-inhibitory molecule CTLA4119; and ectonucleotidases CD39 and CD73 to deplete pro-inflammatory extracellular ATP.118 These diverse mechanisms contribute to the immunosuppressive functions of Treg cells in self and tumor tolerance.
To balance effector responses, Treg cells undergo adaptation in the periphery. Compared with naive T cells, Treg cells display an antigen-experienced, activated phenotype due to their TCR-mediated recognition of self-tissue and also require IL-2 signaling and mTORC1-mediated anabolic metabolism for functional fitness.118,120 Further, in response to specific cytokine and inflammatory cues, Treg cells co-opt the transcriptional and trafficking programs of effector T cells, resembling plastic effector responses (Figure 3A). For instance, under Th1-prone conditions, Treg cells acquire T-bet and CXCR3 expression and Th1-like programming to counteract T-bet-expressing CD4+ and CD8+ T cells.121–123 Further, follicular Treg cells express Bcl6 and CXCR5 to downmodulate Tfh cell activity and germinal center reactions.124,125 The plasticity of Treg cells ensures a precise spatiotemporal control of the context-specific effector responses while limiting immunopathology.118
Although Treg cells are beneficial for mediating protection from autoimmunity, restraining effector responses may be detrimental in certain conditions. In particular, intratumoral Treg cell accumulation is a key barrier to antitumor immunity and immunotherapy and correlates with decreased patient survival126 (Figure 3C). Intratumoral Treg cells are heterogeneous, and only the FOXP3hi, but not FOXP3lo, subpopulation shows immunosuppressive capabilities in patients with colorectal cancer.127 Additionally, Th1-like Treg cells accumulate in tumors and co-localize with CXCL9-producing cDC1s to suppress CD8+ T cell function.128 However, intratumoral Treg cell adaptation can result in aberrant IFNγ expression that reprograms the tumor microenvironment to be more inflammatory, thereby boosting antitumor immunity.129 Intratumoral Treg cells show unique requirements for uptake of nutrients such as lactic acid130 and lipids,131 as well as de novo lipid synthesis,132 and depend upon CTLA4 to maintain metabolic homeostasis and lineage stability.133 Strategies for therapeutic targeting of Treg cells for tumor therapy include their depletion and functional reprogramming by modulating co-inhibitory receptor (e.g., CTLA4), cytokine (e.g., IL-2), and chemokine receptor (e.g., CCR4 and CCR8) signaling.126 However, immune-associated adverse events remain a challenge for Treg cell-targeted cancer immunotherapies, with future therapeutics likely benefiting from a better understanding of tumor context-specific functional programming. Additional studies are also warranted to ascertain the functional contributions of other immunosuppressive populations (e.g., Tr1 cells,134 KIR+ CD8+ T cells,135 and regulatory B cells136) to immune health and disease, as their interplay with Treg cells is likely essential for balancing pro-inflammatory and immunosuppressive responses to different insults.
HOW DOES THE IMMUNE SYSTEM BALANCE TISSUE IMMUNITY AND HOMEOSTASIS?
In a successful acute immune response, the offending agent is cleared, inflammation resolved, and homeostasis re-established. Importantly, a primary immune response leaves the previously infected tissue more capable of defense against a subsequent infection. Resident memory lymphocytes enter non-lymphoid tissues (though some also populate lymphoid tissues), where they perceive and respond to environmental cues and are maintained for long periods of time.137 They defend against future pathogen encounters and prevent the dangerous pathology associated with innate immune defenses triggered during a primary response. An inability to return to normal homeostatic set points results in chronic inflammation and multiple diseases, including allergic disease, fibrosis, autoimmunity, and neurodegenerative disorders, and such effects are counteracted by various intratissue and inter-tissue interactions between lymphocytes and tissue cells and factors. In this section, we discuss the diverse types of tissue lymphocytes and define how these “sentinels” contribute to defense against infection while supporting tissue function and organismal homeostasis.
Memory tissue lymphocytes are lifelong sentinels
Antigen-experienced lymphocytes form both circulating and TRM populations. Antigen-specific TRM cells alter the rules of engagement for subsequent immune responses to the same invading agent, as these highly specialized cells clear pathogens at an exponentially faster rate than most innate mechanisms.137,138 Within hours of re-infection, TRM cells recruit, localize, and activate innate immune cells in tissues, a process that takes up to a day in their absence.139 TRM and tissue-resident B (BRM) cells are anatomically, transcriptionally, epigenetically, and functionally distinct from their circulating counterparts.137,140–142 Although CD8+ TRM cells are well studied, there is also a growing interest in the more recently described CD4+ TRM and BRM cells.
Resident memory cells are optimized to clear offending agents through their interactions with both the previously encountered offending agent and the tissue environment in which they need to functionally engage. A two-site continuum of lymphocyte spatiotemporal differentiation occurs as the immune response progresses: lymphocytes are primed to gain functional properties in the lymphoid tissues and then learn how to communicate with their neighbors and serve as sentinels upon tissue entry. In both sites, differentiation processes occur such that broad classes of lymphocytes contribute to the memory pool: some that are more terminally differentiated with capacity to exert strong effector functions and others that serve as a more plastic, proliferative reserve population that can repopulate the effector-like pool.140,143 Below, we discuss characteristics of lymphocytes that populate the tissues to provide protection against pathogens while also regulating homeostasis and tissue repair (Figure 4).
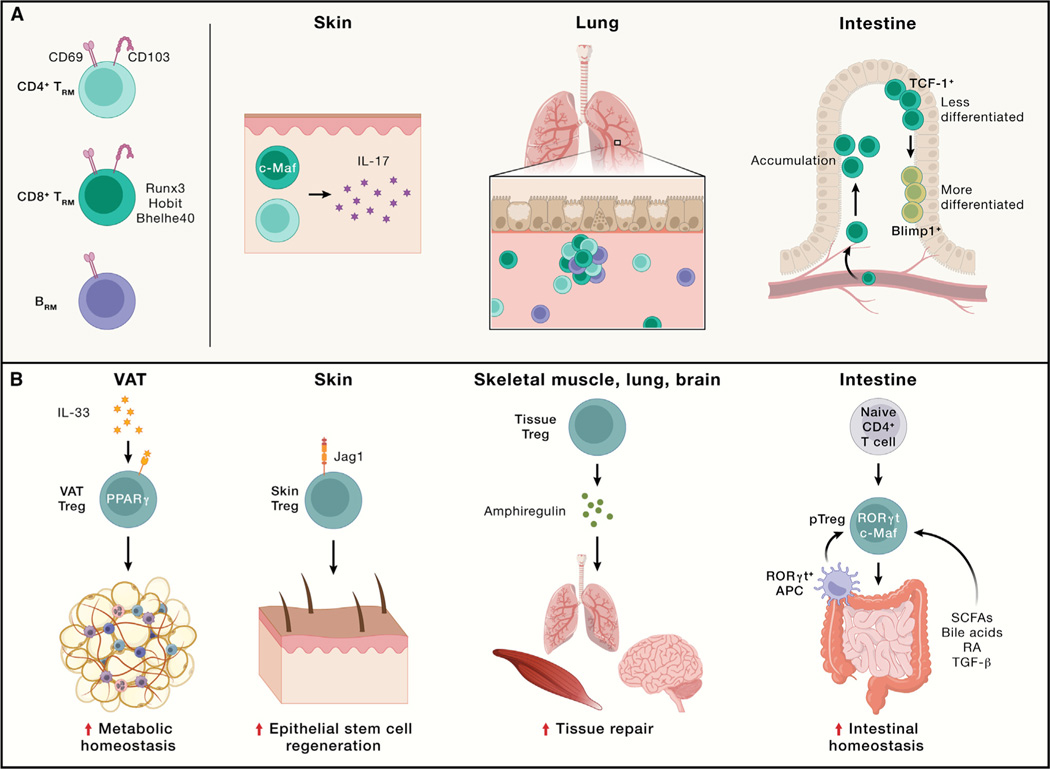
Figure 4. Functional importance of lymphocytes in non-lymphoid tissues in maintaining tissue immunity and homeostasis.
(A) TRM and BRM cells in different non-lymphoid tissues or tumors. CD69 is a typical tissue-resident marker for TRM and BRM cells, while CD103 expression is more limited in lineage and tissue location compared with CD69. CD8+ TRM cells are more extensively studied, with transcriptional regulators such as Runx3, Hobit, and Bhelhe40 identified in certain tissues. In the skin, both CD4+ and CD8+ TRM cells producing IL-17 are present, with CD8+ TRM cells requiring c-Maf but not Runx3 for their generation. In the lung, CD4+ TRM, CD8+ TRM, and BRM cells are present during influenza infection. In the intestine, two distinct subsets of CD8+ TRM cells are present: one with enhanced memory potential and high TCF-1 expression, while the other expresses high levels of Blimp1.
(B).Treg cells in different non-lymphoid tissues. Visceral adipose tissue (VAT) Treg cells, regulated by PPARγ and IL-33 signaling, orchestrate metabolic homeostasis and downmodulate tissue inflammation. In the skin, Treg cells expressing Notch ligand Jagged 1 (Jag1) localize to hair follicles and promote epithelial stem cell regeneration. Upon tissue injury, Treg cells produce amphiregulin to promote tissue repair in multiple tissues, including the skeletal muscle, lung, and brain. In the intestine, peripheral Treg (pTreg) cells are derived from naive T cells in response to short-chain fatty acids (SCFAs), secondary bile acids, retinoic acid (RA), or TGF-β and are marked by RORγt and c-Maf expression. RORγt+ APCs are crucial for intestinal pTreg cell generation.
Functional diversity of resident memory lymphocytes
CD8+ TRM cells are critical for imparting local immune protection against different infections.137 Moreover, murine CD8+ TRM cells accelerate immune control of viral, bacterial, fungal, and parasitic infections.137 In humans, TRM cells are present in virtually every tissue examined. Transcriptional profiling of human T cells from various tissue sites revealed a conserved transcriptional program that is distinct from blood memory T cells but shares key gene expression profiles with mouse TRM cells.141 In humans, the frequency of antigen-specific TRM cells correlates with better control of viruses, including hepatitis B virus in the liver,144 HIV in the lymphoid tissue,145 and RSV in the lung.146
Although most CD8+ TRM cells share some core signatures associated with migration and adhesion, there is also significant heterogeneity associated with the type of immunological insult that primes the response and the subsequent site of TRM residency.147 Because of this heterogeneity, diverse transcriptional and metabolic programs regulate the formation of each unique TRM population. For example, deletion of the transcription factor Runx3 has varying effects on IFNγ-expressing CD8+ TRM numbers in distinct non-lymphoid tissues,148 and skin-resident CD8+ TRM cells that produce IL-17 upon bacterial infection and wound healing require c-Maf but not Runx3 for their generation.149 Intratissue heterogeneity even exists among antigen-specific TRM cells. For instance, CD8+ TRM cells within a tissue express varying levels of key TRM-associated molecules (e.g., CD69, CD103, IL18R, TCF-1, and T-bet), and the frequency of cells expressing these molecules can shift over time.147 In the small intestine, two distinct subsets of TRM cells are observed: one with enhanced memory potential that expresses high levels of Id3 and TCF-1 but low levels of Blimp1, while the other expresses high levels of Blimp1 but low levels of Id3.140 Of interest, CD8+ tumor-infiltrating lymphocytes (TILs) can also display characteristics of TRM cells in certain mouse and human cancers, which usually correlate with better antitumor immunity.74,150,151 Understanding the distinctions between these subsets and how to properly regulate their distribution and function may therefore be pertinent to enhancing protective immunity against both infection and tumors.
Although CD8+ TRM cells may be optimally poised to kill pathogens at the site of infection, CD4+ TRM cells are the great communicators of the system. Like CD4+ T cells in a primary immune response, CD4+ TRM cells interact with many cell types through ligand-receptor interactions and secrete a diverse array of cytokines. Moreover, functionally diverse CD4+ TRM populations contribute to the control of infections with viruses,152–155 bacteria,156 and parasites.157 CD4+ TRM cells share transcriptional programs with CD8+ TRM cells and depend upon IL-2 signaling in the lymphoid tissue to become tissue resident.153,158 CD4+ TRM cells can also undergo further diversification within the tissues, with some retaining a more effector-like phenotype159 and others behaving more like Tfh cells.160,161 Whether this division of labor is a characteristic of only T-bet-expressing Th1-like TRM cells during influenza infection162 or is also a general feature of CD4+ TRM cells responding to other types of immunologic insults requires further investigation.
BRM cells can also protect against re-infection.163–165 Recent studies using fluorescently labeled probes have characterized influenza-specific memory B cells and begun to shed light on the phenotype, longevity, functional attributes, and interactions with other tissue-resident immune populations.142,143 BRM cells are randomly distributed throughout the lung, near local alveoli.143 Upon re-infection, these activated BRM cells rapidly migrate to sites of infection, where they produce antibodies to help clear the virus. A survey of mouse and human tissues characterized Bmem cells in the lung and gut tissues that express both shared and unique phenotypic markers, which will aid our understanding of BRM cells.166
Tissue Treg cells support tissue homeostasis and repair
The optimal functionality of TRM cells can be a double-edged sword by providing immune protection and contributing to immunopathology. For instance, CD4+ TRM cells can promote allergic and autoimmune diseases of the skin, lung, nervous system, and gut.137 As such, Treg cells contribute to counteracting these effects by promoting tissue homeostasis, as well as regeneration and repair in response to various insults.118 Treg cells in non-lymphoid tissues, such as those in the visceral adipose tissue (VAT), adapt to specific microenvironmental signals,167 and accordingly, VAT Treg cells have distinct transcriptome profiles (partly mediated by PPARγ168 and IL-33169) and TCR repertoires than splenic Treg cells. These VAT Treg cells orchestrate tissue homeostasis via local and systemic regulation of metabolism, partly by downmodulating tissue inflammation that may predispose to type 2 diabetes and metabolic syndromes,167 and their generation requires a stepwise, multi-site differentiation process.170 Beyond VAT, scRNA-seq profiling reveals tissue-specific signatures of Treg cells and their progressive adaptation to tissue sites under steady state.171,172 Tissue Treg cells also promote tissue regeneration via interfacing with stem cells. For example, in the skin, Treg cells localize to hair follicles, where they stimulate stem cell function or regeneration via Notch signaling.173 In a mouse model of vitiligo, the proper position and function of skin Treg cells are mediated by CCR5 signaling, which likely extends to human vitiligo patients.174 Finally, in response to tissue-specific insults, Treg cells support tissue repair and physiological function by production of growth factors.118 In particular, upon tissue injury, Treg cells produce amphiregulin to promote tissue repair and regeneration in the skeletal muscle,175 lung,176,177 and brain.178 In a mouse model of influenza infection, Treg cell-derived amphiregulin prevents lung tissue damage without affecting antiviral immune responses, revealing discrete functions of Treg cells in mediating immune suppression and tissue repair.176 The extent to which Treg cells interplay with tissue cells and impact physiological function and tissue repair, and the underlying processes, are fruitful areas of future studies.
Commensal microbes interplay with the immune system in barrier tissues, and specialized tissue Treg cells called peripheral Treg (pTreg) cells prevent immune reactivity to microbiota to preserve tissue homeostasis. Upon antigen stimulation, pTreg cells are derived from naive T cells in response to commensal microbe-derived signals (e.g., short-chain fatty acids, secondary bile acids, and retinoic acid) or TGF-β.179 In the intestines, these pTreg cells are marked by RORγt and c-Maf expression. Exposure to food antigens also induces pTreg cell differentiation.179 RORγt+ APCs are crucial for intestinal pTreg cell generation, in part, via integrin αVβ8 expression that enables TGF-β activation.180–182 The identification of the interactions between RORγt+ APCs and pTreg cells reveals an important mechanism for peripheral tolerance that distinguishes commensal microbes from invading harmful insults.180–182 The prevalence and functionality of pTreg cells and their corresponding APCs in other tissues warrant further investigation.
Where do lymphocytes fit into organismal homeostasis?
Organismal homeostasis maintains the internal stability of the host while adjusting for changing external conditions, including alterations in temperature, exposure to toxins or infectious agents, or dietary changes. Sensing of these external insults by the immune system not only activates direct defense mechanisms, but if life is perceived to be threatened, also engages both local and system-wide changes to clear the offending agent. In a physiological emergency (e.g., infection with a replicating pathogen and injury or tissue stress), the immune system overrides the normal set points of homeostasis to help control the infection or heal the injury, a process referred to as inflammation.183 The immune system also contributes to the re-establishment of organismal homeostasis through interactions with the other systems of the body. As discussed below, communication between the immune and nervous systems creates a complex homeostatic network that synergistically enhances the individual function of each system. Although innate lymphoid cells (ILCs) contribute to sensing and responding to perturbations to homeostasis in an antigen-independent manner,184 recent studies have identified similar types of interactions between lymphocytes and the nervous system that depend upon sensing of a specific antigen, with major implications for therapeutic interventions.
Lymphocytes and neurons communicate using a common language of cytokines, hormones, neuropeptides, chemokines, and their receptors,185 and neuroimmune interactions are observed in the central and peripheral nervous systems. Recent evidence has highlighted the ability of antigen-specific lymphocytes to direct neuronal activity in multiple tissue sites. In the skin dermis, IL-17-producing CD4+ TRM cells that are specific for commensal micro-biota co-localize with sensory nerve fibers, where they promote axonal growth and nerve regeneration upon injury.186 In the intestine, sensitization to food allergens drives antigen-specific avoidance behavior that depends upon B cell-secreted IgE, suggesting that antibodies may expand the sensory capacity of the nervous system.187 Remarkably, this allergen-specific response persists for at least 48 weeks after allergic sensitization, further suggesting a specific role for gut-resident memory lymphocytes. However, these additive responses are not always protective. For example, IL-5Rα-expressing nociceptive nerves in the lung are activated by IL-5 produced by ILC2s and Th2 cells, creating a feedforward loop that enhances immunopathology.188
The peripheral nervous system also modulates immune responses in lymphoid and non-lymphoid organs. In response to fasting, catecholaminergic neurons are activated in the ventrolateral medulla, which can shunt CD4+ and CD8+ T cells from the blood and secondary lymphoid organs into the bone marrow in a CXCR4-dependent manner.189,190 Catecholamines contribute to CD8+ T cell exhaustion in both chronic viral infection and cancer, further providing evidence of the intersection of neuronal regulation of the tissue immune response. Specifically, exhausted antigen-specific CD8+ T cells are in close proximity to sympathetic nerves, and ablation of β-adrenergic signaling synergizes with ICB to improve T cell effector function, revealing a potential new therapeutic modality.191 Together, these studies highlight the importance of understanding neuroimmune interactions in tissues and in the larger context of how the body responds to homeostatic perturbations across all systems.
HOW ARE DISTINCT IMMUNE RESPONSES REGULATED?
Adaptive immune cells employ similar and unique strategies against different threats, orchestrated by extrinsic and intrinsic molecular processes that vary across space and time (Figure 5). The extrinsic signals from antigens, co-stimulation, and cytokines (signals 1–3) are predominantly delivered from activated DCs. Nutrients (signal 4) and other cellular interactions also regulate lymphocyte specialization. These extrinsic cues are integrated by cell-intrinsic signaling pathways, metabolism, and epigenetic programs, which culminate in altered gene transcription and protein translation instructing cell state and fate.
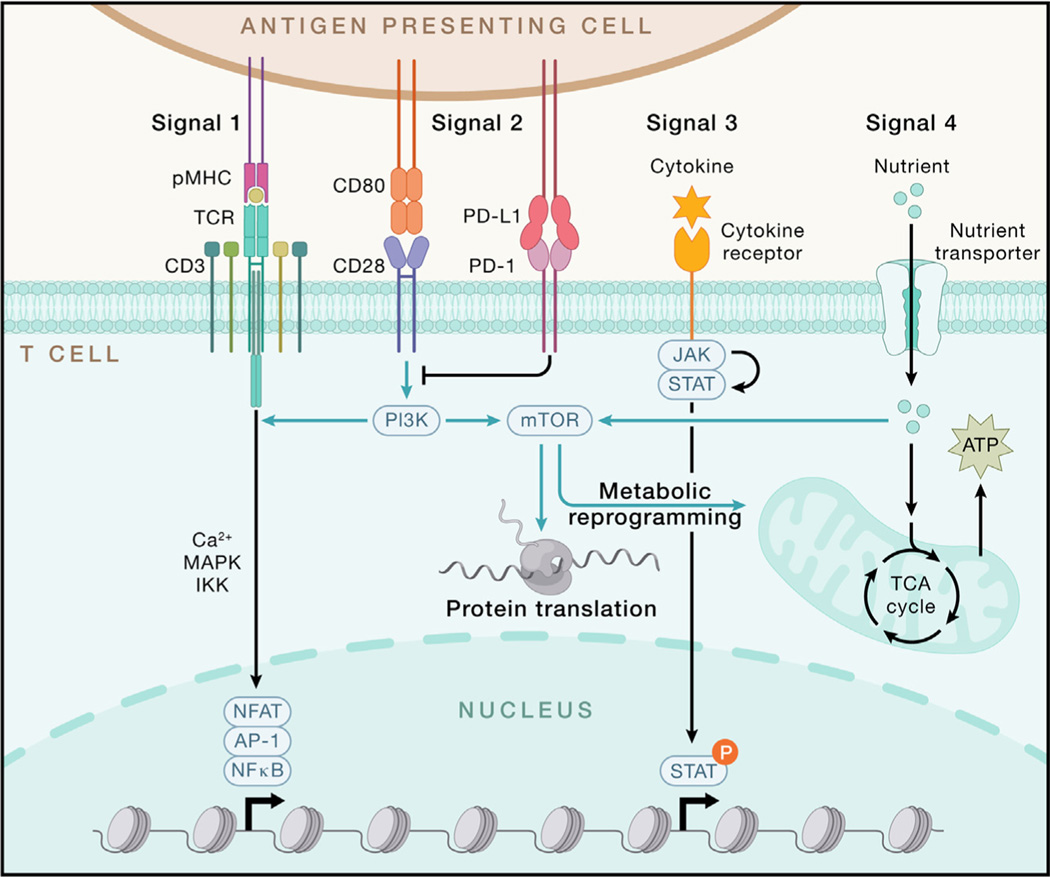
Figure 5. Integration of extrinsic signals and intrinsic programs drives T cell responses.
T cell activation largely relies on signals 1–3 (antigen, co-stimulation, and cytokines). TCR stimulation occurs upon recognition of the peptide presented by MHC molecule (pMHC), which is expressed on antigen-presenting cells (APCs). TCR-pMHC engagement initiates downstream signaling, leading to the activation of transcription factors NFAT, AP-1, and NF-κB mainly through Ca2+, mitogen-activated protein kinase (MAPK), and IKK-dependent signaling, respectively. CD28 serves as the major signal 2 for T cell activation, which induces downstream PI3K-AKT-mTOR signaling, while PD-1 delivers a co-inhibitory signal to block CD28-dependent signaling. Upon binding their receptors, signal 3 cytokines activate JAK-induced STAT phosphorylation, which is pivotal for potentiating T cell activation and differentiation. Nutrients function as signal 4 to license T cell immunity partly through fueling the tricarboxylic acid (TCA) cycle to generate ATP and activating mTOR to enhance metabolism and protein translation during T cell activation. P, phosphorylation.
Extrinsic signals drive adaptive immunity
The diverse stimuli of adaptive immunity largely converge on signals 1–3, which mediate the specificity, strength, and durability of immune responses (Figure 5). During naive T cell activation, TCR stimulation (signal 1) occurs upon recognition of DC-presented pMHC, leading to immunological synapse formation, which shapes TCR signaling192 and mechanical force.193 Downstream of antigen receptors, graded IRF4 expression establishes the fate and function of CD8+ T cells,194,195 CD4+ T cells,196 and B cells.197 Acute and persistent antigen stimulation promote the generation of effector and exhausted T cells, respectively.91 Of note, pMHC-TCR interaction is among the most complex ligand-receptor interaction systems for directing specific biological responses, associated with the vast diversity of antigens and antigen receptors as described above.
However, TCR signaling is not sufficient to elicit a productive adaptive immune response, and this serves to prevent spurious T cell activation and uncontrolled inflammation. Instead, co-stimulatory (signal 2) and cytokine (signal 3) signals act in coordination with antigen stimulation to instruct adaptive immunity. For example, CD28 (signal 2) and IL-12 (signal 3) help promote naive T cell activation. Other co-stimulatory (e.g., ICOS) and co-inhibitory signals (e.g., PD-1, CTLA4, LAG3, TIM-3, and TIGIT) also serve as respective “accelerators” and “brakes” for signal 2. For signal 3, immunostimulatory (e.g., IL-1, IL-2, and IL-6) and immunosuppressive (e.g., TGF-β and IL-10) cytokines shape T cell fate. Importantly, the interplay between signals 1–3 orchestrates adaptive immune responses, including by activating metabolic rewiring (e.g., via TCR and CD28)198,199 or tuning downstream signaling (e.g., ICOS-mediated phosphatidylinositol 3-kinase [PI3K] activation for Tfh responses200). Also, PD-1-targeted ICB therapies enhance CD28-dependent effector function and accumulation of CD8+ T cells in chronic infection and cancer and also act in synergy with IL-2 signaling.91,201–203 How these co-stimulatory and cytokine signals are delivered by specific innate stimuli and cells, including “non-traditional” APCs (e.g., epithelial cells), as well as the impact of other non-traditional signals (e.g., serum amyloid A proteins derived from intestinal epithelial cells that act in collaboration with Th17 cell-promoting cytokines204), will be important to explore.
Nutrients and metabolites license T cell immunity by functioning as signal 4,205,206 including acting on intratumoral CD8+ T cells.91 In particular, glucose and glutamine are limiting for intratumoral lymphocytes due to the enhanced capacities of intratumoral myeloid cells and tumors cells to acquire and consume these nutrients.207 Consequently, intratumoral glucose restriction impairs T cell activation and function.208,209 Similarly, glutamine shapes metabolic reprogramming210 and context-specific function211,212 of T cells, as well as intratumoral cDC1s.213 Intratumoral glutamine administration rectifies defective cDC1-dependent CD8+ T cell antitumor immunity and overcomes therapeutic resistance to immunotherapies in animal models.213 Other amino acids, such as arginine and methionine, are also required for T cell function,214,215 partly by orchestrating mTORC1 activation,205,216 whereas uptake of lactic acid217 and oxidized lipids218 is largely detrimental to intratumoral T cell function. Finally, G-protein-coupled receptors orchestrate T cell immunity,219 partly by mediating signals from stress-associated hormones (e.g., catecholamines)191 or the dietary nutrient trans-vaccenic acid.220 Future research should explore how adaptive immunity is shaped by nutrient availability and usage in various tissues or from dietary and microbiota-derived sources.
Cell-intrinsic programs
How do lymphocytes interpret and integrate these diverse signals? Antigen and co-stimulatory receptor engagement induces a series of phosphorylation events and other post-translational modifications, leading to the activation of key transcription factors nuclear factor κB (NF-κB) (via IKK signaling), AP-1 (via ERK signaling), and NFAT (via calcium-calcineurin signaling). Additionally, cytokines primarily activate the JAK-STAT pathway, while nutrient-dependent signaling is mediated by a three-tier process composed of nutrient transporters, sensors, and transducers.205 As with signals 1–4 themselves, crosstalk and integration of such signaling pathways regulate adaptive immunity, as illustrated by the cooperation between NFAT and AP-1221 or STAT and lineage-specific transcription factors, thereby leading to transcriptional activation of signature cytokines and effector molecules. Feedback control mechanisms also exist to prevent T cell hyperactivation, including those mediated by the Cbl family proteins222 and Regnase-1.223 In particular, emerging studies highlight the important roles of metabolic and epigenetic rewiring that shape gene transcription and cell fate choices.
Metabolic regulation of adaptive immunity has been revealed over the past decade.15 Glycolysis promotes effector over Treg cell differentiation, and fatty acid metabolism contributes to memory T cell generation.68,224,225 Also, mitochondrial dysregulation is a hallmark of exhausted CD8+ T cells.226–228 Mechanistically, extensive crosstalk between immune signaling and metabolic programs directs “bidirectional metabolic signaling.”15 For instance, mTORC1 integrates signals 1–4 and shows reciprocal interplay with Myc, thereby driving anabolic metabolism, quiescence exit, and T cell differentiation.199,210 Beyond generating ATP, mitochondria are signaling hubs in T cell biology, as specific mitochondrial metabolites (e.g., α-ketoglutarate229 and the oncometabolite d-2-hydroxyglutarate230) impact the epigenetic landscape or protein activity in T cells. Moreover, new metabolic regulators of T cell fate, including phosphatidylethanolamine and guanosine diphosphate (GDP)-fucose signaling, which respectively drive Tfh and terminal effector CD8+ T cell differentiation, were identified by CRISPR-based genetic screens.231,232 Altogether, cell state or fate is characterized by unique metabolic profiles, nutrient requirements, and tissue-specific regulators. Metabolic adaptation represents a key means to alter the fitness and function of adaptive immune cells to the specific immuno-logical context.
Epigenetic remodeling confers phenotypic stability of immune cells and is more reflective of cell fate alterations than transcriptional and metabolic events. As such, epigenetic analyses can resolve key questions in adaptive immune responses. For instance, chromatin state analyses show that exhausted CD8+ T cells represent a separate T cell lineage in chronic infection233–235 and cancer.236,237 Chromatin accessibility analyses also revealed the progressive differentiation of exhausted T cells,236,238 while global mapping of histone methylation in effector CD4+ T cells exposes subset-specific patterns of signature cytokines and plasticity for master transcription factors.239 Accordingly, targeting the DNA methyltransferase Dnmt3a alters CD8+ T cell differentiation in acute and chronic responses,80,240 while disruption of the chromatin remodeling complex SWI/SNF affects CD8+ T cell differentiation241–244 and Treg cell activation and function.205,245 These findings offer new opportunities for therapeutic targeting of epigenetic programs for cancer and immune-mediated diseases.
ENGINEERING IMMUNITY: VACCINES AND THERAPEUTICS
The hallmarks of T cell and B cell responses (cytotoxic and helper functions and antibody secretion) are the core components mediating effective and durable protection in the context of vaccines. Further, these hallmarks are leveraged as tools for therapeutic interventions, including the use of T cell-based adoptive cell therapies (ACTs) as “living drugs” and monoclonal antibodies (mAbs). Harnessing the power and unique features of the adaptive immune system holds great promise to treat a broad range of diseases, which is briefly described below mainly from the perspectives of vaccination for infectious diseases and engineered therapies for cancer.
Vaccination as a means to understand and utilize adaptive immunity
Vaccination is, in many ways, where the understanding of adaptive immunity began, with the Jennerian cowpox vaccine for smallpox.246 Vaccines aim to generate protective immune memory that will aid clearance and limit the pathology and inflammation of an infectious challenge. The majority of vaccines are benchmarked on their ability to induce serum antibody responses, which are generally strong correlates of protection for most vaccines.247 While animal models and human studies have reported correlations between vaccine efficacy and T cell response features, including magnitude and quality in various systems, there are no rigorously validated, epidemiologically established T cell-based correlates of protection for any vaccine in clinical use. This is likely because assays for T cell enumeration and characterization are more time consuming, expensive, and difficult to compare than antibody assays. Although few studies have directly compared the predictive power of antibodies versus cellular responses in human patients, the contributions of CD4+ and CD8+ T cells versus B cells can be defined mechanistically with genetic ablation and depletion studies in animal models. The recent interest of defining correlates of protection to COVID-19 vaccines has highlighted the utility of animal models for determining the precise mechanisms of protection and the need for improved human assays that can assess multiple arms of immunity.
The understanding of human vaccinology has advanced significantly during the COVID-19 pandemic.248,249 One striking finding concerned the length of germinal center reactions after mRNA vaccination against COVID-19 that has revolutionized modern vaccine development. In mouse studies, most germinal center reactions induced by immunization are resolved within 6 weeks; however, in humans, naive COVID-19 mRNA vaccine responses for both Tfh and B cells are still detected at 6 months after boosting, when the response is peaking in many individuals.250–252 The kinetics of short-lived plasmablasts in the blood and of bone marrow-resident, long-lived plasma cells were also carefully measured.253 The rapid dynamic decay of the plasma-blast response corresponds to measured declines in serum antibody.254 However, paired with the knowledge of ongoing germinal center function and output (as measured by increased numbers of Bmem cells255), the maturation of improved and higher affinity antibodies is still occurring, despite waning total antibody levels.256 The rapid evolution of SARS-CoV-2 led to escape from the neutralizing antibody responses raised against the ancestral vaccine antigens. However, CD4+ and CD8+ T cell responses are well characterized for these vaccines, and there is strong evidence that T cells play an important role in limiting serious infection-associated pathology.249 These data were derived from both animal studies and from humans who were on treatments that suppressed the B cell response.257 A greater understanding of the quantitative contribution of T cell responses to human vaccine protection will aid future vaccine design programs.
Inducing potent immune memory by vaccination requires a balance between activating robust lymphocyte responses and causing inflammatory sequelae. This balancing act can be achieved by adjuvants, “the immunologist’s dirty little secret” described by Charles Janeway before mechanisms of action for vaccines were understood.258 In modern vaccines, adjuvants that activate a single innate immune pathway (e.g., specific Toll-like receptors) are being tested for their capacity to prime a proper adaptive immune response.259 Further, modern vaccine design also focuses on driving specific forms of adaptive immune memory. For example, the generation of both CD4+ and CD8+ TRM cells is a major priority of next-generation vaccine initiatives, with mucosal vaccine platforms, such as inactivated viruses and inhaled vaccines in the airways, being tested.260 The goal is to localize lymphocyte memory into the upper or lower airways to more faithfully mirror the potent TRM responses generated by a pathogenic infection of the respiratory system. However, as with pathogenic infection, vaccine-induced inflammation in the airways, lung, intestinal, or genital mucosa via a vaccination needs to be carefully regulated to limit any immunopathology.
Beyond infectious diseases, there are rapidly growing efforts in vaccination against tumor antigens, including unmutated tumor-associated antigens and personalized or public neoantigens.261 For most tumor vaccination, the immunization occurs after the tumor has formed and seeks to elicit cytotoxic CD4+ and CD8+ T cell responses against the tumor. Immunizing against tumor antigens presents complications due to the close or identical relationship of the target antigen to self-antigens and the unforgiving environment of the tumor and local lymphatics. As such, the tumor microenvironment can limit T cell activity by suppressing function and limiting migration. However, recent tumor vaccination efforts have shown significant promise in solid tumors, including in pancreatic cancer and melanoma.262 In addition, certain tumors are promoted by viral infection, including the human papillomavirus (HPV), which plays a causative role in the development of many cervical and head and neck cancers. The introduction of HPV vaccines has profoundly lowered the risk of cancer in recipient populations, demonstrating the potential efficacy for preventive anti-cancer vaccines for tumors where there are established, shared antigens.263 In combination with other immunotherapeutic approaches, tumor vaccines are an important area for future mechanistic investigation.
T cells as living drugs
T cell-based cell therapies are built upon decades of basic research. The convergence of immunobiology and synthetic biology (called synthetic immunology) has broadened the applicability of cell therapies, especially by pioneering the use of genetically reprogrammed T cells to treat tumors, infectious disease, and autoimmunity.264 Cellular therapies for cancer include use of T cells that are expanded after isolation from the tumor site (TILs), chimeric antigen receptor T cells (CAR-T), and TCR-engineered T cells (TCR-T) (Figure 6A). TIL therapy has shown promising efficacy265 and was approved by the US Food and Drug Administration (FDA) in 2024 to treat melanoma (Amtagvi). However, isolation and preparation of abundant tumor-specific T cells are still difficult in other cancer types. CARs are fusion proteins that combine the antigen-binding domains of antibodies with T cell signaling domains264 and recognize cancer cell surface antigens independently of MHC-mediated peptide presentation. CAR-T therapy has achieved strong clinical outcomes in treating B cell malignancies,266–268 leading to FDA approval of CD19-targeting CARs to treat B cell leukemias and lymphomas. By contrast, CAR-T cells have shown limited efficacy toward solid tumors, whose immunosuppressive tumor microenvironment often causes CAR-T cell exclusion and exhaustion. Also, it remains challenging to identify target antigens with high-level and homogenous expression on solid tumors but not normal tissues. Logic-gated CAR-T cells, which express two different CARs on their surface and must sense two separate antigens for activation,269,270 may overcome this obstacle. Finally, TCR-T therapy uses an ectopically expressed TCR recognizing a tumor antigen to expand tumor-specific T cells for enhanced killing activity. TCR-T cells are subject to MHC restriction and thus will only function in individuals with that MHC.264
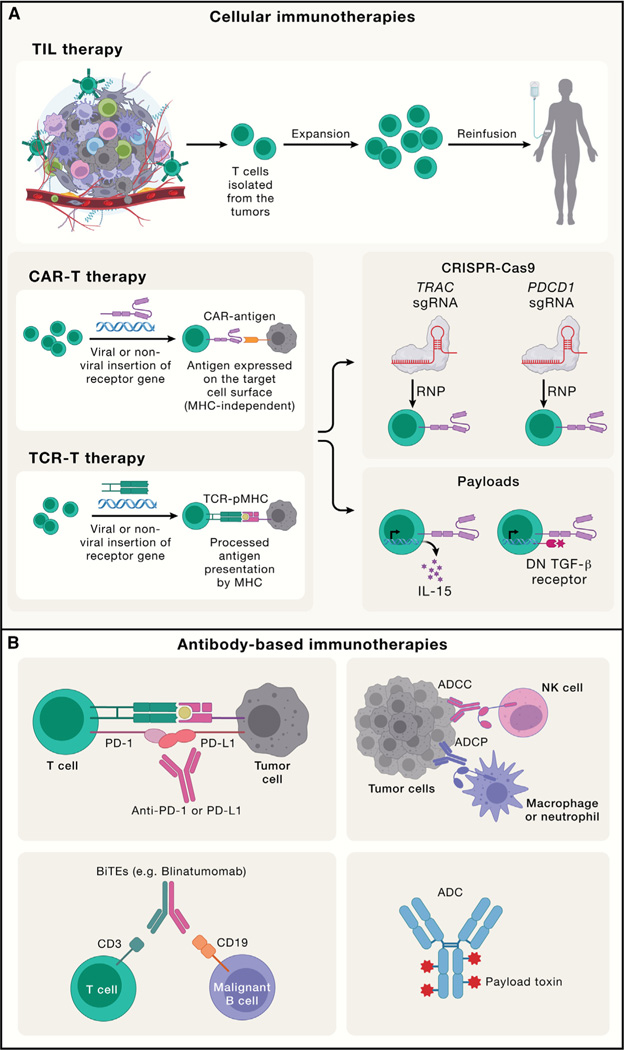
Figure 6. Engineering adaptive immune responses for cell-and antibody-based therapies.
(A).TIL, CAR-T, and TCR-T are three prominent cellular immunotherapies for cancer. For TIL therapy, T cells are isolated from the tumors for expansion and reinfusion into tumor patients; for CAR-T cells, CARs are engineered to recognize surface antigen independently of MHC-mediated antigen presentation; for TCR-T, a specific TCR gene recognizing the tumor antigen is ectopically expressed in T cells to expand tumor-specific T cells for tumor killing, which requires peptide-MHC (pMHC) expression on tumor cells. For CAR-T or TCR-T therapies, new strategies have been developed to modify intracellular signaling pathways in these tumor-specific T cells to enhance tumor killing, such as CRISPR-mediated genome editing of the TRAC or PDCD1 locus or introduction of payloads (e.g., cytokines like IL-15 and dominant-negative [DN] TGF-β receptor) expressed on T cells. These strategies increase the antitumor function in these cellular therapies. RNP, Cas9-expressing ribonucleoprotein.
(B).Antibody-based immunotherapies include ICB targeting co-inhibitory molecules (e.g., PD-1 or PD-L1), which unleash endogenous T cell-mediated antitumor responses; antibody-dependent cellular cytotoxicity (ADCC; mainly mediated by NK cells) or antibody-dependent cellular phagocytosis (ADCP; mainly mediated by macrophages and neutrophils); a BiTE that bridges T cells and tumor cells by targeting the CD3 chain of TCRs on the T cells and CD19 on malignant B cells to promote T cell-mediated tumor killing; and ADCs, which are composed of a tumor antigen-targeting antibody, a linker, and a payload toxin.
New strategies to modify intracellular metabolic, proliferation, and survival pathways can further boost the efficacies of these cell therapies. Moreover, CRISPR technology has advanced genetic engineering to improve T cell potency by offering applications such as unbiased functional screens and site-specific genetic modifications.271 Recent in vivo or multimodal CRISPR screens in T cells identified new targets that, when deleted, reprogram cells to boost CAR-T cell function.271–274 In vitro CRISPR screens uncovered that IFNγR signaling is required for CAR binding duration and avidity in solid but not liquid tumors.275 Also, inserting CAR expression into specific genetic loci (e.g., TRAC or PDCD1) through CRISPR technology further improves CAR-T efficiency.276,277 Finally, engineered T cells can act as vehicles to deliver therapeutic “payloads” to improve effector function. These payloads include cytokines (e.g., IL-15278 and IL-10279 to enhance persistence and resistance to dysfunction, respectively), dominant-negative receptors as a “sink” for immunosuppressive cytokines (e.g., TGF-β280), or antibodies (e.g., anti-PD-1 to overcome exhaustion281). Beyond cancer, T cell-based cell therapies are emerging for infectious disease and other immune-mediated diseases, such as engineering of CAR-Treg cells to treat autoimmune diseases and transplantation.264 Opportunities and challenges remain for these living drugs in the future for these non-malignant diseases.
Antibodies as therapies
Due to their high specificity for target antigens, mAbs represent a major category of therapies for cancer, inflammation, and other diseases (Figure 6B). Following the approval of the first anti-CD3 mAb therapy for transplant rejection (muromonab, more commonly called OKT3) in 1986, antibody-based therapies have rapidly emerged with remarkable clinical and commercial success. Further, mAb engineering has evolved from the first generation (mouse antibodies with names ending in -omab) to the second (human-mouse chimera, -ximab), the third (humanized, -zumab), and finally the fourth (fully human, -umab) generation, each associated with diverse mechanisms of action.
By targeting co-inhibitory molecules such as CTLA4 and PD-1 (or its ligand PD-L1) via therapeutic antibodies, ICB therapy directly unleashes endogenous T cell responses to target cancer.282 Though many patients experience durable tumor regression, most patients are unresponsive or develop resistance to current forms of ICB. Predicting the response to ICB remains challenging, although the composition or gene expression profiles of immune cells in tumors and the periphery serve as the most pertinent markers.282 Beyond the cancer context, ICB immunotherapies are being tested to treat malaria and HIV infection.283 Other antibodies that block ligand-receptor interactions are highly efficacious in the clinic, as exemplified by blocking cytokines (e.g., TNF-α by adalimumab,284 IL-4Rα by dupilumab,285 and IL-17 by secukinumab67) that have transformed the treatment of autoimmune and allergic diseases.
Besides targeting ligand-receptor interactions, these therapeutic antibodies direct cell-mediated cytotoxicity toward tumors. For instance, anti-CD20 mAbs may induce caspase-independent programmed cell death of malignant B cells,286 whereas antibodies targeting death receptors cause caspase-dependent apoptosis.287 Moreover, antibodies elicit cell-mediated immune reactions, whereby other immune cells (namely, natural killer [NK] cells and phagocytes) target and kill antigen-expressing target cells bound by the therapeutic antibodies. Compared with mAbs using full-length IgG antibodies, the bispecific T cell engager (BiTE) is composed of an antibody fragment for activating the TCR-CD3 chain and a tumor-specific antigen to bridge the interaction between T cells and target cancer cells, thereby inducing an artificial immune synapse that promotes T cell-mediated killing of target cells independent of TCR specificity.288 Finally, there is a growing interest in antibody-drug conjugates (ADCs) composed of a tumor antigen-targeting antibody, a linker, and a payload toxin (tubulin-binding or DNA-targeting agents). After the ADC is internalized by target tumor cells, the linker is cleaved to release the toxin.289 Improving ADC-related efficacy while reducing side effects requires further optimization.
Aside from the direct use of T cells and antibodies as therapies, cytokine-based therapies, such as small-molecule inhibitors of JAK, have achieved clinical success in autoimmunity and inflammation.290 Whereas direct use of cytokines has shown limited effects, possibly due to their pleiotropic effects and rapid diffusion, innovations in bioengineering have recently yielded promising new technologies in modulating adaptive immunity. For example, although IL-2 drives both expansion and exhaustion of CD8+ T cells, a partial agonist of IL-2 retains the ability to induce expansion and better maintain the TCF-1+ stem-like phenotype to escape exhaustion.291 Overall, limited efficacy remains a major hurdle of these immune-based therapies, and combination therapies are key to overcoming therapeutic resistance.
CONCLUDING REMARKS
Here, we have highlighted the core and emerging principles of adaptive immunity and its applications to immunotherapy. Since the discoveries of T cells and B cells in the 1960s, exploring adaptive immunity has yielded pivotal contributions to immunology. Adaptive immune cells are also an excellent system to study fundamentals of cell, molecular, and structural biology and mechanisms of metabolic, signaling, epigenetic, and transcriptional regulation, leading to major discoveries beyond immunology. For instance, investigating immunometabolism at the signaling, cellular, and systemic (e.g., dietary interventions and microbiota-derived nutrients) levels has enriched our knowledge on immunity and metabolism and provided new opportunities for targeting cancer, metabolic disorders, and other immune-mediated diseases. Similarly, investigation of epigenetic and chromatin regulation in lymphocytes has generated new concepts in context-specific gene regulation. Continued exploration of adaptive immunity and its intersection with fundamental biology will advance our understanding of the principles that are unique to the immune system and those that are broadly applicable to other biological systems.
Tissue immunity is an emerging regulator of physiology and disease. Unlike other cells, adaptive immune cells face the unique challenges of entering and acclimating to varying environmental cues in diverse tissues. Tissue context-specific adaptation is therefore fundamental to understanding tissue physiology and disease pathogenesis; however, much remains to be learned about the impact of the diverse tissue microenvironments on the adaptation of immune cells, and vice versa, as well as the complex immune-tissue cell crosstalk. To that end, recent interrogations of adaptive immunity in non-lymphoid tissues have markedly advanced our knowledge of tissue immunity (e.g., tissue-resident cells and Treg-mediated tissue repair). More importantly, the integration of adaptive immunity with physiology leads to an in-depth understanding of normal and diseased states of various tissues and the organism. For instance, the information on tumor-immune or neuroimmune interactions has contributed to therapeutic interventions of cancer and other diseases. However, we are only beginning to understand how the immune system shapes tissue homeostasis or dysregulation in aging-related, neurodegenerative, or metabolic disorders. How adaptive immunity, including host-intrinsic (e.g., tissue-specific immunity) and extrinsic (e.g., infection, microbiota, or nutrition) factors, intersects with normal physiology and disease states will likely bring fundamental new insights into immunobiology, with remarkable translational and therapeutic potentials.
Immunology is at the forefront of technology development and innovation, with ever-increasing resolution and ever-expanding dimensions to probe the complex adaptive immune system. Historically, the application of flow cytometry permitted single-cell analysis of immune responses, which was instrumental for discovery and analysis of lymphocyte subsets, whereas mouse conditional genetic models allowed for cell type-specific or spatiotemporally controlled modulation of gene function in vivo. Recent innovative technologies have provided unprecedented opportunities for high-throughput analysis of adaptive immune responses. For instance, the use of scRNA-seq permits the measurement of thousands of transcripts rather than limited markers offered by flow cytometry and can be integrated with protein expressional data and spatially resolved information (e.g., via spatial transcriptomics). Further, CRISPR-based pooled screens in vivo enable the unbiased discovery of the most functionally relevant targets under physiologically relevant conditions. Similarly, we have moved from analysis of individual antigen receptors to unbiased profiling of the entire TCR or BCR repertoire at single-cell resolution. The integrative use of these multi-omics tools requires expertise in computational biology and systems biology to uncover biological insights and disease targets. Therefore, systems immunology serves as a primary example of successful integration of experimental biology and data science for biological and clinical discovery. The synthesis of experimental and computational biology and integration of immunobiology, cutting-edge technologies, and data science will drive the next-generation research in adaptive immunity and produce new innovations.
Adaptive immunity may also hold the key to understanding and curing a broad spectrum of human diseases. This notion is best exemplified by the successes in vaccine development for infectious disease, immunotherapies for cancer, and cytokine-targeting therapies for inflammatory diseases. Indeed, basic research and fundamental discoveries using animal models (e.g., immune checkpoint molecules, inflammatory cytokines, and Th17 cells) have laid the foundation for innovative immunotherapies and clinical translation. Aside from this traditional bench-to-bedside translation, bedside-to-bench is being increasingly applied. In particular, with the advances in high-throughput technologies (e.g., scRNA-seq, spatial transcriptomics, and multiplexed imaging), the use of human patient-derived materials has moved beyond observational phenotyping to in-depth mechanistic investigation of disease state and precision medicine, accelerated by complementary model systems, such as organoids and advanced mouse genetic models (e.g., humanized mice). Moreover, immune engineering, as evidenced by the remarkable success of CAR-T living drugs and antibody and cytokine engineering, will continue to drive innovation and new opportunities for translation. In summary, the intersection of adaptive immunity with biology, physiology, technology, and therapy has produced major breakthroughs and transformed biological and clinical sciences, and we are undoubtedly on the brink of many more breakthroughs in the next half century.
ACKNOWLEDGMENTS
The authors acknowledge N. Chapman and H. Shi in the Chi Laboratory for insightful scientific discussions and editing of the manuscript. We apologize to our colleagues whose work is not highlighted due to space constraints. Research support is provided by the National Institutes of Health (NIH) (grants CA253188, CA281868, AI105887, AI131703, AI140761, AI150241, and AI150514), the Lupus Research Alliance, and ALSAC (to H.C.); AI165457, AI177874, AI142001, and AI176545 (to M.P.); and AI150747, AI154470, AI144616, AI136514, CA265009, HHS contract no. 75N93019C00052 (CIVR-HRP), 75N93021C00016 (SJCEIRR), and 75N93021C00018 (CIDER) (to P.G.T.).
Footnotes
DECLARATION OF INTERESTS
H.C. has consulted for Kumquat Biosciences, Inc., Chugai Pharmaceutical Co., and ONO Pharmaceutical Co. and is a co-inventor on patents/patent applications in the fields of immunotherapy. M.P. serves on the scientific advisory board of Vaxart, Inc. P.G.T. has consulted or received honoraria and/or travel support from Illumina, J&J, Pfizer, Merck, and 10x Genomics, and his lab has received funding from ElevateBio. P.G.T. serves on the scientific advisory boards of ImmunoScape, Shennon Bio, and CytoAgents.
REFERENCES
- 1.Cooper MD (2015). The early history of B cells. Nat. Rev. Immunol. 15, 191–197. 10.1038/nri3801. [DOI] [PubMed] [Google Scholar]
- 2.Miller JFAP (2020). The function of the thymus and its impact on modern medicine. Science 369, eaba2429. 10.1126/science.aba2429. [DOI] [PubMed] [Google Scholar]
- 3.Lu LL, Suscovich TJ, Fortune SM, and Alter G. (2018). Beyond binding: antibody effector functions in infectious diseases. Nat. Rev. Immunol. 18, 46–61. 10.1038/nri.2017.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dupic T, Marcou Q, Walczak AM, and Mora T. (2019). Genesis of the αβ T-cell receptor. PLoS Comput. Biol. 15, e1006874. 10.1371/journal.pcbi.1006874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Farber DL, Netea MG, Radbruch A, Rajewsky K, and Zinkernagel RM (2016). Immunological memory: lessons from the past and a look to the future. Nat. Rev. Immunol. 16, 124–128. 10.1038/nri.2016.13. [DOI] [PubMed] [Google Scholar]
- 6.Belz GT, Smith CM, Kleinert L, Reading P, Brooks A, Shortman K, Carbone FR, and Heath WR (2004). Distinct migrating and nonmigrating dendritic cell populations are involved in MHC class I-restricted antigen presentation after lung infection with virus. Proc. Natl. Acad. Sci. USA 101, 8670–8675. 10.1073/pnas.0402644101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meuer SC, Schlossman SF, and Reinherz EL (1982). Clonal analysis of human cytotoxic T lymphocytes: T4+ and T8+ effector T cells recognize products of different major histocompatibility complex regions. Proc. Natl. Acad. Sci. USA 79, 4395–4399. 10.1073/pnas.79.14.4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bjorkman PJ, Saper MA, Samraoui B, Bennett WS, Strominger JL, and Wiley DC (1987). Structure of the human class I histocompatibility antigen, HLA-A2. Nature 329, 506–512. 10.1038/329506a0. [DOI] [PubMed] [Google Scholar]
- 9.Doherty PC, and Zinkernagel RM (1975). H-2 compatibility is required for T-cell-mediated lysis of target cells infected with lymphocytic choriomeningitis virus. J. Exp. Med. 141, 502–507. 10.1084/jem.141.2.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lythe G, Callard RE, Hoare RL, and Molina-Parίs C. (2016). How many TCR clonotypes does a body maintain? J. Theor. Biol. 389, 214–224. 10.1016/j.jtbi.2015.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu T, Guan J, Handel A, Tscharke DC, Sidney J, Sette A, Wakim LM, Sng XYX, Thomas PG, Croft NP, et al. (2019). Quantification of epitope abundance reveals the effect of direct and cross-presentation on influenza CTL responses. Nat. Commun. 10, 2846. 10.1038/s41467-019-10661-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moon JJ, Chu HH, Pepper M, McSorley SJ, Jameson SC, Kedl RM, and Jenkins MK (2007). Naive CD4(+) T cell frequency varies for different epitopes and predicts repertoire diversity and response magnitude. Immunity 27, 203–213. 10.1016/j.immuni.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Obar JJ, Khanna KM, and Lefrançois L. (2008). Endogenous naive CD8+ T cell precursor frequency regulates primary and memory responses to infection. Immunity 28, 859–869. 10.1016/j.immuni.2008.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaud G, Lesourne R, and Love PE (2018). Regulatory mechanisms in T cell receptor signalling. Nat. Rev. Immunol. 18, 485–497. 10.1038/s41577-018-0020-8. [DOI] [PubMed] [Google Scholar]
- 15.Chapman NM, Boothby MR, and Chi H. (2020). Metabolic coordination of T cell quiescence and activation. Nat. Rev. Immunol. 20, 55–70. 10.1038/s41577-019-0203-y. [DOI] [PubMed] [Google Scholar]
- 16.Heinzel S, Binh Giang T, Kan A, Marchingo JM, Lye BK, Corcoran LM, and Hodgkin PD (2017). A Myc-dependent division timer complements a cell-death timer to regulate T cell and B cell responses. Nat. Immunol. 18, 96–103. 10.1038/ni.3598. [DOI] [PubMed] [Google Scholar]
- 17.De Boer RJ, Homann D, and Perelson AS (2003). Different dynamics of CD4+ and CD8+ T cell responses during and after acute lymphocytic choriomeningitis virus infection. J. Immunol. 171, 3928–3935. 10.4049/jimmunol.171.8.3928. [DOI] [PubMed] [Google Scholar]
- 18.Yoon H, Kim TS, and Braciale TJ (2010). The cell cycle time of CD8+ T cells responding in vivo is controlled by the type of antigenic stimulus. PLoS One 5, e15423. 10.1371/journal.pone.0015423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wells AD, Gudmundsdottir H, and Turka LA (1997). Following the fate of individual T cells throughout activation and clonal expansion. Signals from T cell receptor and CD28 differentially regulate the induction and duration of a proliferative response. J. Clin. Invest. 100, 3173–3183. 10.1172/JCI119873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaech SM, Wherry EJ, and Ahmed R. (2002). Effector and memory T-cell differentiation: implications for vaccine development. Nat. Rev. Immunol. 2, 251–262. 10.1038/nri778. [DOI] [PubMed] [Google Scholar]
- 21.Halle S, Halle O, and Förster R. (2017). Mechanisms and Dynamics of T Cell-Mediated Cytotoxicity In Vivo. Trends Immunol. 38, 432–443. 10.1016/j.it.2017.04.002. [DOI] [PubMed] [Google Scholar]
- 22.Kohlmeier JE, Cookenham T, Miller SC, Roberts AD, Christensen JP, Thomsen AR, and Woodland DL (2009). CXCR3 directs antigen-specific effector CD4+ T cell migration to the lung during parainfluenza virus infection. J. Immunol. 183, 4378–4384. 10.4049/jimmunol.0902022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nolz JC, Starbeck-Miller GR, and Harty JT (2011). Naive, effector and memory CD8 T-cell trafficking: parallels and distinctions. Immuno-therapy 3, 1223–1233. 10.2217/imt.11.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Masopust D, and Schenkel JM (2013). The integration of T cell migration, differentiation and function. Nat. Rev. Immunol. 13, 309–320. 10.1038/nri3442. [DOI] [PubMed] [Google Scholar]
- 25.Cyster JG, and Allen CDC (2019). B Cell Responses: Cell Interaction Dynamics and Decisions. Cell 177, 524–540. 10.1016/j.cell.2019.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heesters BA, van der Poel CE, Das A, and Carroll MC (2016). Antigen Presentation to B Cells. Trends Immunol. 37, 844–854. 10.1016/j.it.2016.10.003. [DOI] [PubMed] [Google Scholar]
- 27.Kuchen S, Robbins R, Sims GP, Sheng C, Phillips TM, Lipsky PE, and Ettinger R. (2007). Essential role of IL-21 in B cell activation, expansion, and plasma cell generation during CD4+ T cell-B cell collaboration. J. Immunol. 179, 5886–5896. 10.4049/jimmunol.179.9.5886. [DOI] [PubMed] [Google Scholar]
- 28.Gonzalez DG, Cote CM, Patel JR, Smith CB, Zhang Y, Nickerson KM, Zhang T, Kerfoot SM, and Haberman AM (2018). Nonredundant Roles of IL-21 and IL-4 in the Phased Initiation of Germinal Center B Cells and Subsequent Self-Renewal Transitions. J. Immunol. 201, 3569–3579. 10.4049/jimmunol.1500497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Young C, and Brink R. (2021). The unique biology of germinal center B cells. Immunity 54, 1652–1664. 10.1016/j.immuni.2021.07.015. [DOI] [PubMed] [Google Scholar]
- 30.Feng Y, Seija N, Di Noia JM, and Martin A. (2020). AID in Antibody Diversification: There and Back Again. Trends Immunol. 41, 586–600. 10.1016/j.it.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ripperger TJ, and Bhattacharya D. (2021). Transcriptional and Metabolic Control of Memory B Cells and Plasma Cells. Annu. Rev. Immunol. 39, 345–368. 10.1146/annurev-immunol-093019-125603. [DOI] [PubMed] [Google Scholar]
- 32.Inoue T, and Kurosaki T. (2024). Memory B cells. Nat. Rev. Immunol.24, 5–17. 10.1038/s41577-023-00897-3. [DOI] [PubMed] [Google Scholar]
- 33.Muroyama Y, and Wherry EJ (2021). Memory T-Cell Heterogeneity and Terminology. Cold Spring Harb. Perspect. Biol. 13, a037929. 10.1101/cshperspect.a037929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bassing CH, Swat W, and Alt FW (2002). The mechanism and regulation of chromosomal V(D)J recombination. Cell 109 (Suppl), S45–S55. 10.1016/s0092-8674(02)00675-x. [DOI] [PubMed] [Google Scholar]
- 35.Mora T, and Walczak A. (2018). Quantifying lymphocyte receptor diversity. In Systems Immunology: An Introduction to Modeling Methods for Scientists, Das J, ed. (CRC Press, Taylor and Francis; ), pp. 1–10. [Google Scholar]
- 36.Robins HS, Campregher PV, Srivastava SK, Wacher A, Turtle CJ, Kahsai O, Riddell SR, Warren EH, and Carlson CS (2009). Comprehensive assessment of T-cell receptor beta-chain diversity in alphabeta T cells. Blood 114, 4099–4107. 10.1182/blood-2009-04-217604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Elhanati Y, Sethna Z, Callan CG Jr., Mora T, and Walczak AM (2018). Predicting the spectrum of TCR repertoire sharing with a data-driven model of recombination. Immunol. Rev. 284, 167–179. 10.1111/imr.12665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Elhanati Y, Murugan A, Callan CG Jr., Mora T, and Walczak AM (2014). Quantifying selection in immune receptor repertoires. Proc. Natl. Acad. Sci. USA 111, 9875–9880. 10.1073/pnas.1409572111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marcou Q, Mora T, and Walczak AM (2018). High-throughput immune repertoire analysis with IGoR. Nat. Commun. 9, 561. 10.1038/s41467-018-02832-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sethna Z, Elhanati Y, Callan CG, Walczak AM, and Mora T. (2019). OLGA: fast computation of generation probabilities of B- and T-cell receptor amino acid sequences and motifs. Bioinformatics 35, 2974–2981. 10.1093/bioinformatics/btz035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Davidsen K, Olson BJ, DeWitt WS 3rd, Feng J, Harkins E, Bradley P, and Matsen F.A.t. (2019). Deep generative models for T cell receptor protein sequences. eLife 8, e46935. 10.7554/eLife.46935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Teng G, Maman Y, Resch W, Kim M, Yamane A, Qian J, Kieffer-Kwon KR, Mandal M, Ji Y, Meffre E, et al. (2015). RAG Represents a Widespread Threat to the Lymphocyte Genome. Cell 162, 751–765. 10.1016/j.cell.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Onozawa M, and Aplan PD (2012). Illegitimate V(D)J recombination involving nonantigen receptor loci in lymphoid malignancy. Genes Chromosomes Cancer 51, 525–535. 10.1002/gcc.21942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Braun TP, Eide CA, and Druker BJ (2020). Response and Resistance to BCR-ABL1-Targeted Therapies. Cancer Cell 37, 530–542. 10.1016/j.ccell.2020.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aplan PD, Lombardi DP, Ginsberg AM, Cossman J, Bertness VL, and Kirsch IR (1990). Disruption of the human SCL locus by “illegitimate” V-(D)-J recombinase activity. Science 250, 1426–1429. 10.1126/science.2255914. [DOI] [PubMed] [Google Scholar]
- 46.Yancopoulos GD, and Alt FW (1985). Developmentally controlled and tissue-specific expression of unrearranged VH gene segments. Cell 40, 271–281. 10.1016/0092-8674(85)90141-2. [DOI] [PubMed] [Google Scholar]
- 47.Barajas-Mora EM, Kleiman E, Xu J, Carrico NC, Lu H, Oltz EM, Murre C, and Feeney AJ (2019). A B-Cell-Specific Enhancer Orchetrates Nuclear Architecture to Generate a Diverse Antigen Receptor Repertoire. Mol. Cell 73, 48–60.e5. 10.1016/j.molcel.2018.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Karki S, Banerjee S, McLean K, Dinner A, and Clark MR (2019). Transcription factories in Igkappa allelic choice and diversity. Adv. Immunol. 141, 33–49. 10.1016/bs.ai.2018.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hudson D, Fernandes RA, Basham M, Ogg G, and Koohy H. (2023). Can we predict T cell specificity with digital biology and machine learning? Nat. Rev. Immunol. 23, 511–521. 10.1038/s41577-023-00835-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Glanville J, Huang H, Nau A, Hatton O, Wagar LE, Rubelt F, Ji X, Han A, Krams SM, Pettus C, et al. (2017). Identifying specificity groups in the T cell receptor repertoire. Nature 547, 94–98. 10.1038/nature22976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dash P, Fiore-Gartland AJ, Hertz T, Wang GC, Sharma S, Souquette A, Crawford JC, Clemens EB, Nguyen THO, Kedzierska K, et al. (2017). Quantifiable predictive features define epitope-specific T cell receptor repertoires. Nature 547, 89–93. 10.1038/nature22383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thomas N, Best K, Cinelli M, Reich-Zeliger S, Gal H, Shifrut E, Madi A, Friedman N, Shawe-Taylor J, and Chain B. (2014). Tracking global changes induced in the CD4 T-cell receptor repertoire by immunization with a complex antigen using short stretches of CDR3 protein sequence. Bioinformatics 30, 3181–3188. 10.1093/bioinformatics/btu523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moris P, De Pauw J, Postovskaya A, Gielis S, De Neuter N, Bittremieux W, Ogunjimi B, Laukens K, and Meysman P. (2021). Current challenges for unseen-epitope TCR interaction prediction and a new perspective derived from image classification. Brief. Bioinform. 22, bbaa318. 10.1093/bib/bbaa318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mayer-Blackwell K, Schattgen S, Cohen-Lavi L, Crawford JC, Souquette A, Gaevert JA, Hertz T, Thomas PG, Bradley P, and Fiore-Gartland A. (2021). TCR meta-clonotypes for biomarker discovery with tcrdist3 enabled identification of public, HLA-restricted clusters of SARS-CoV-2 TCRs. eLife 10, e68605. 10.7554/eLife.68605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shugay M, Bagaev DV, Zvyagin IV, Vroomans RM, Crawford JC,Dolton G, Komech EA, Sycheva AL, Koneva AE, Egorov ES, et al. (2018). VDJdb: a curated database of T-cell receptor sequences with known antigen specificity. Nucleic Acids Res. 46, D419–D427. 10.1093/nar/gkx760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sewell AK (2012). Why must T cells be cross-reactive? Nat. Rev. Immunol. 12, 669–677. 10.1038/nri3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Klein L, Kyewski B, Allen PM, and Hogquist KA (2014). Positive and negative selection of the T cell repertoire: what thymocytes see (and don’t see). Nat. Rev. Immunol. 14, 377–391. 10.1038/nri3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Michelson DA, and Mathis D. (2022). Thymic mimetic cells: tolerogenic masqueraders. Trends Immunol. 43, 782–791. 10.1016/j.it.2022.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nemazee D. (2017). Mechanisms of central tolerance for B cells. Nat. Rev. Immunol. 17, 281–294. 10.1038/nri.2017.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dong C. (2021). Cytokine Regulation and Function in T Cells. Annu. Rev. Immunol. 39, 51–76. 10.1146/annurev-immunol-061020-053702. [DOI] [PubMed] [Google Scholar]
- 61.Hirota K, Duarte JH, Veldhoen M, Hornsby E, Li Y, Cua DJ, Ahlfors H, Wilhelm C, Tolaini M, Menzel U, et al. (2011). Fate mapping of IL-17-producing T cells in inflammatory responses. Nat. Immunol. 12, 255–263. 10.1038/ni.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Harbour SN, Maynard CL, Zindl CL, Schoeb TR, and Weaver CT (2015). Th17 cells give rise to Th1 cells that are required for the pathogenesis of colitis. Proc. Natl. Acad. Sci. USA 112, 7061–7066. 10.1073/pnas.1415675112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Karmaus PWF, Chen X, Lim SA, Herrada AA, Nguyen TM, Xu B, Dhungana Y, Rankin S, Chen W, Rosencrance C, et al. (2019). Metabolic heterogeneity underlies reciprocal fates of TH17 cell stemness and plasticity. Nature 565, 101–105. 10.1038/s41586-018-0806-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gagliani N, Amezcua Vesely MC, Iseppon A, Brockmann L, Xu H, Palm NW, de Zoete MR, Licona-Limón P, Paiva RS, Ching T, et al. (2015). Th17 cells transdifferentiate into regulatory T cells during resolution of inflammation. Nature 523, 221–225. 10.1038/nature14452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Panzer M, Sitte S, Wirth S, Drexler I, Sparwasser T, and Voehringer D. (2012). Rapid in vivo conversion of effector T cells into Th2 cells during helminth infection. J. Immunol. 188, 615–623. 10.4049/jimmunol.1101164. [DOI] [PubMed] [Google Scholar]
- 66.Wang YH, Voo KS, Liu B, Chen CY, Uygungil B, Spoede W, Bernstein JA, Huston DP, and Liu YJ (2010). A novel subset of CD4(+) T(H)2 memory/effector cells that produce inflammatory IL-17 cytokine and promote the exacerbation of chronic allergic asthma.J. Exp. Med. 207, 2479–2491. 10.1084/jem.20101376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schnell A, Littman DR, and Kuchroo VK (2023). TH17 cell heterogeneity and its role in tissue inflammation. Nat. Immunol. 24, 19–29. 10.1038/s41590-022-01387-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shi LZ, Wang R, Huang G, Vogel P, Neale G, Green DR, and Chi H. (2011). HIF1alpha-dependent glycolytic pathway orchestrates a metabolic checkpoint for the differentiation of TH17 and Treg cells. J. Exp. Med. 208, 1367–1376. 10.1084/jem.20110278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wu L, Hollinshead KER, Hao Y, Au C, Kroehling L, Ng C, Lin WY, Li D, Silva HM, Shin J, et al. (2020). Niche-Selective Inhibition of Pathogenic Th17 Cells by Targeting Metabolic Redundancy. Cell 182, 641–654.e20. 10.1016/j.cell.2020.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang C, Yosef N, Gaublomme J, Wu C, Lee Y, Clish CB, Kaminski J, Xiao S, Meyer Zu Horste G, Pawlak M, et al. (2015). CD5L/AIM Regulates Lipid Biosynthesis and Restrains Th17 Cell Pathogenicity. Cell 163, 1413–1427. 10.1016/j.cell.2015.10.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Puleston DJ, Baixauli F, Sanin DE, Edwards-Hicks J, Villa M, Kabat AM, Kamiński MM, Stanckzak M, Weiss HJ, Grzes KM, et al. (2021). Polyamine metabolism is a central determinant of helper T cell lineage fidelity. Cell 184, 4186–4202.e20. 10.1016/j.cell.2021.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wagner A, Wang C, Fessler J, DeTomaso D, Avila-Pacheco J, Kaminski J, Zaghouani S, Christian E, Thakore P, Schellhaass B, et al. (2021). Metabolic modeling of single Th17 cells reveals regulators of autoimmunity. Cell 184, 4168–4185.e21. 10.1016/j.cell.2021.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Song W, and Craft J. (2023). T Follicular Helper Cell Heterogeneity. Annu. Rev. Immunol. 10.1146/annurev-immunol-090222-102834. [DOI] [PubMed] [Google Scholar]
- 74.Zheng L, Qin S, Si W, Wang A, Xing B, Gao R, Ren X, Wang L, Wu X, Zhang J, et al. (2021). Pan-cancer single-cell landscape of tumor-infiltrating T cells. Science 374, abe6474. 10.1126/science.abe6474. [DOI] [PubMed] [Google Scholar]
- 75.Helmink BA, Reddy SM, Gao J, Zhang S, Basar R, Thakur R, Yizhak K, Sade-Feldman M, Blando J, Han G, et al. (2020). B cells and tertiary lymphoid structures promote immunotherapy response. Nature 577, 549–555. 10.1038/s41586-019-1922-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang F, Wei K, Slowikowski K, Fonseka CY, Rao DA, Kelly S, Goodman SM, Tabechian D, Hughes LB, Salomon-Escoto K, et al. (2019). Defining inflammatory cell states in rheumatoid arthritis joint synovial tissues by integrating single-cell transcriptomics and mass cytometry. Nat. Immunol. 20, 928–942. 10.1038/s41590-019-0378-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Meckiff BJ, Ramıŕez-Suástegui C, Fajardo V, Chee SJ, Kusnadi A, Simon H, Eschweiler S, Grifoni A, Pelosi E, Weiskopf D, et al. (2020). Imbalance of Regulatory and Cytotoxic SARS-CoV-2-Reactive CD4(+) T Cells in COVID-19. Cell 183, 1340–1353.e16. 10.1016/j.cell.2020.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gate D, Saligrama N, Leventhal O, Yang AC, Unger MS, Middeldorp J, Chen K, Lehallier B, Channappa D, De Los Santos MB, et al. (2020). Clonally expanded CD8 T cells patrol the cerebrospinal fluid in Alzheimer’s disease. Nature 577, 399–404. 10.1038/s41586-019-1895-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Oh DY, Kwek SS, Raju SS, Li T, McCarthy E, Chow E, Aran D, Ilano A, Pai CS, Rancan C, et al. (2020). Intratumoral CD4+ T Cells Mediate Anti-tumor Cytotoxicity in Human Bladder Cancer. Cell 181, 1612–1625.e13. 10.1016/j.cell.2020.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Youngblood B, Hale JS, Kissick HT, Ahn E, Xu X, Wieland A, Araki K, West EE, Ghoneim HE, Fan Y, et al. (2017). Effector CD8 T cells dedifferentiate into long-lived memory cells. Nature 552, 404–409. 10.1038/nature25144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pais Ferreira D, Silva JG, Wyss T, Fuertes Marraco SA, Scarpellino L, Charmoy M, Maas R, Siddiqui I, Tang L, Joyce JA, et al. (2020). Central memory CD8+ T cells derive from stem-like Tcf7hi effector cells in the absence of cytotoxic differentiation. Immunity 53, 985–1000.e11. 10.1016/j.immuni.2020.09.005. [DOI] [PubMed] [Google Scholar]
- 82.Johnnidis JB, Muroyama Y, Ngiow SF, Chen Z, Manne S, Cai Z, Song S, Platt JM, Schenkel JM, Abdel-Hakeem M, et al. (2021). Inhibitory signaling sustains a distinct early memory CD8+ T cell precursor that is resistant to DNA damage. Sci. Immunol. 6, eabe3702. 10.1126/sciimmunol.abe3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bresser K, Kok L, Swain AC, King LA, Jacobs L, Weber TS, Perié L, Duffy KR, de Boer RJ, Scheeren FA, and Schumacher TN (2022). Replicative history marks transcriptional and functional disparity in the CD8+ T cell memory pool. Nat. Immunol. 23, 791–801. 10.1038/s41590-022-01171-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chang JT, Palanivel VR, Kinjyo I, Schambach F, Intlekofer AM, Banerjee A, Longworth SA, Vinup KE, Mrass P, Oliaro J, et al. (2007). Asymmetric T lymphocyte division in the initiation of adaptive immune responses. Science 315, 1687–1691. 10.1126/science.1139393. [DOI] [PubMed] [Google Scholar]
- 85.Jeannet G, Boudousquié C, Gardiol N, Kang J, Huelsken J, and Held W. (2010). Essential role of the Wnt pathway effector Tcf-1 for the establishment of functional CD8 T cell memory. Proc. Natl. Acad. Sci. USA 107, 9777–9782. 10.1073/pnas.0914127107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhou X, Yu S, Zhao DM, Harty JT, Badovinac VP, and Xue HH (2010). Differentiation and persistence of memory CD8(+) T cells depend on T cell factor 1. Immunity 33, 229–240. 10.1016/j.immuni.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gautam S, Fioravanti J, Zhu W, Le Gall JB, Brohawn P, Lacey NE, Hu J, Hocker JD, Hawk NV, Kapoor V, et al. (2019). The transcription factor c-Myb regulates CD8+ T cell stemness and antitumor immunity. Nat. Immunol. 20, 337–349. 10.1038/s41590-018-0311-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hess Michelini R, Doedens AL, Goldrath AW, and Hedrick SM (2013). Differentiation of CD8 memory T cells depends on Foxo1.J. Exp. Med. 210, 1189–1200. 10.1084/jem.20130392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Buck MD, O’Sullivan D, Klein Geltink RI, Curtis JD, Chang CH, Sanin DE, Qiu J, Kretz O, Braas D, van der Windt GJ, et al. (2016). Mitochondrial Dynamics Controls T Cell Fate through Metabolic Programming. Cell 166, 63–76. 10.1016/j.cell.2016.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zajac AJ, Blattman JN, Murali-Krishna K, Sourdive DJ, Suresh M, Altman JD, and Ahmed R. (1998). Viral immune evasion due to persistence of activated T cells without effector function. J. Exp. Med. 188, 2205–2213. 10.1084/jem.188.12.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Giles JR, Globig AM, Kaech SM, and Wherry EJ (2023). CD8+ T cells in the cancer-immunity cycle. Immunity 56, 2231–2253. 10.1016/j.immuni.2023.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Seo H, Chen J, González-Avalos E, Samaniego-Castruita D, Das A, Wang YH, López-Moyado IF, Georges RO, Zhang W, Onodera A, et al. (2019). TOX and TOX2 transcription factors cooperate with NR4A transcription factors to impose CD8+ T cell exhaustion. Proc. Natl. Acad. Sci. USA 116, 12410–12415. 10.1073/pnas.1905675116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Scott AC, Dündar F, Zumbo P, Chandran SS, Klebanoff CA, Shakiba M, Trivedi P, Menocal L, Appleby H, Camara S, et al. (2019). TOX is a critical regulator of tumour-specific T cell differentiation. Nature 571, 270–274. 10.1038/s41586-019-1324-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Alfei F, Kanev K, Hofmann M, Wu M, Ghoneim HE, Roelli P, Utzschneider DT, von Hoesslin M, Cullen JG, Fan Y, et al. (2019). TOX reinforces the phenotype and longevity of exhausted T cells in chronic viral infection. Nature 571, 265–269. 10.1038/s41586-019-1326-9. [DOI] [PubMed] [Google Scholar]
- 95.Khan O, Giles JR, McDonald S, Manne S, Ngiow SF, Patel KP, Werner MT, Huang AC, Alexander KA, Wu JE, et al. (2019). TOX transcriptionally and epigenetically programs CD8+ T cell exhaustion. Nature 571, 211–218. 10.1038/s41586-019-1325-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yao C, Sun HW, Lacey NE, Ji Y, Moseman EA, Shih HY, Heuston EF, Kirby M, Anderson S, Cheng J, et al. (2019). Single-cell RNA-seq reveals TOX as a key regulator of CD8+ T cell persistence in chronic infection. Nat. Immunol. 20, 890–901. 10.1038/s41590-019-0403-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Beltra JC, Manne S, Abdel-Hakeem MS, Kurachi M, Giles JR, Chen Z, Casella V, Ngiow SF, Khan O, Huang YJ, et al. (2020). Developmental Relationships of Four Exhausted CD8(+) T Cell Subsets Reveals Underlying Transcriptional and Epigenetic Landscape Control Mechanisms. Immunity 52, 825–841.e8. 10.1016/j.immuni.2020.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Im SJ, Hashimoto M, Gerner MY, Lee J, Kissick HT, Burger MC, Shan Q, Hale JS, Lee J, Nasti TH, et al. (2016). Defining CD8+ T cells that provide the proliferative burst after PD-1 therapy. Nature 537, 417–421. 10.1038/nature19330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.He R, Hou S, Liu C, Zhang A, Bai Q, Han M, Yang Y, Wei G, Shen T, Yang X, et al. (2016). Follicular CXCR5- expressing CD8(+) T cells curtail chronic viral infection. Nature 537, 412–428. 10.1038/nature19317. [DOI] [PubMed] [Google Scholar]
- 100.Utzschneider DT, Charmoy M, Chennupati V, Pousse L, Ferreira DP, Calderon-Copete S, Danilo M, Alfei F, Hofmann M, Wieland D, et al. (2016). T Cell Factor 1-Expressing Memory-like CD8+ T Cells Sustain the Immune Response to Chronic Viral Infections. Immunity 45, 415–427. 10.1016/j.immuni.2016.07.021. [DOI] [PubMed] [Google Scholar]
- 101.Leong YA, Chen Y, Ong HS, Wu D, Man K, Deleage C, Minnich M, Meckiff BJ, Wei Y, Hou Z, et al. (2016). CXCR5+ follicular cytotoxic T cells control viral infection in B cell follicles. Nat. Immunol. 17, 1187–1196. 10.1038/ni.3543. [DOI] [PubMed] [Google Scholar]
- 102.Wu T, Ji Y, Moseman EA, Xu HC, Manglani M, Kirby M, Anderson SM, Handon R, Kenyon E, Elkahloun A, et al. (2016). The TCF1-Bcl6 axis counteracts type I interferon to repress exhaustion and maintain T cell stemness. Sci. Immunol. 1, eaai8593. 10.1126/sciimmunol.aai8593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Miller BC, Sen DR, Al Abosy R, Bi K, Virkud YV, LaFleur MW, Yates KB, Lako A, Felt K, Naik GS, et al. (2019). Subsets of exhausted CD8+ T cells differentially mediate tumor control and respond to checkpoint blockade. Nat. Immunol. 20, 326–336. 10.1038/s41590-019-0312-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Siddiqui I, Schaeuble K, Chennupati V, Fuertes Marraco SA, Calderon-Copete S, Pais Ferreira D, Carmona SJ, Scarpellino L, Gfeller D, Pradervand S, et al. (2019). Intratumoral Tcf1+PD-1+CD8+ T Cells with Stem-like Properties Promote Tumor Control in Response to Vaccination and Checkpoint Blockade Immunotherapy. Immunity 50, 195–211.e10. 10.1016/j.immuni.2018.12.021. [DOI] [PubMed] [Google Scholar]
- 105.Tsui C, Kretschmer L, Rapelius S, Gabriel SS, Chisanga D, Knöpper K, Utzschneider DT, Nüssing S, Liao Y, Mason T, et al. (2022). MYB orchestrates T cell exhaustion and response to checkpoint inhibition. Nature 609, 354–360. 10.1038/s41586-022-05105-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hudson WH, Gensheimer J, Hashimoto M, Wieland A, Valanparambil RM, Li P, Lin JX, Konieczny BT, Im SJ, Freeman GJ, et al. (2019). Proliferating Transitory T Cells with an Effector-like Transcriptional Signature Emerge from PD-1+ Stem-like CD8+ T Cells during Chronic Infection. Immunity 51, 1043–1058.e4. 10.1016/j.immuni.2019.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zander R, Schauder D, Xin G, Nguyen C, Wu X, Zajac A, and Cui W. (2019). CD4+ T Cell Help Is Required for the Formation of a Cytolytic CD8+ T Cell Subset that Protects against Chronic Infection and Cancer. Immunity 51, 1028–1042.e4. 10.1016/j.immuni.2019.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ando S, Perkins CM, Sajiki Y, Chastain C, Valanparambil RM, Wieland A, Hudson WH, Hashimoto M, Ramalingam SS, Freeman GJ, et al. (2023). mTOR regulates T cell exhaustion and PD-1-targeted immunotherapy response during chronic viral infection. J. Clin. Invest. 133, e160025. 10.1172/JCI160025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhou P, Shi H, Huang H, Sun X, Yuan S, Chapman NM, Connelly JP, Lim SA, Saravia J, KC A, et al. (2023). Single-cell CRISPR screens in vivo map T cell fate regulomes in cancer. Nature 624, 154–163. 10.1038/s41586-023-06733-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Huang Q, Wu X, Wang Z, Chen X, Wang L, Lu Y, Xiong D, Liu Q, Tian Y, Lin H, et al. (2022). The primordial differentiation of tumor-specific memory CD8+ T cells as bona fide responders to PD-1/PD-L1 blockade in draining lymph nodes. Cell 185, 4049–4066.e25. 10.1016/j.cell.2022.09.020. [DOI] [PubMed] [Google Scholar]
- 111.Dahling S, Mansilla AM, Knopper K, Grafen A, Utzschneider DT, Ugur M, Whitney PG, Bachem A, Arampatzi P, et al. (2022). Type 1 conventional dendritic cells maintain and guide the differentiation of precursors of exhausted T cells in distinct cellular niches. Immunity 55, 656–670.e8. 10.1016/j.immuni.2022.03.006. [DOI] [PubMed] [Google Scholar]
- 112.Schenkel JM, Herbst RH, Canner D, Li A, Hillman M, Shanahan SL, Gibbons G, Smith OC, Kim JY, Westcott P, et al. (2021). Conventional type I dendritic cells maintain a reservoir of proliferative tumor-antigen specific TCF-1+ CD8+ T cells in tumor-draining lymph nodes. Immunity 54, 2338–2353.e6. 10.1016/j.immuni.2021.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Connolly KA, Kuchroo M, Venkat A, Khatun A, Wang J, William I, Hornick NI, Fitzgerald BL, Damo M, Kasmani MY, et al. (2021). A reservoir of stem-like CD8+ T cells in the tumor-draining lymph node preserves the ongoing antitumor immune response. Sci. Immunol. 6, eabg7836. 10.1126/sciimmunol.abg7836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Rahim MK, Okholm TLH, Jones KB, McCarthy EE, Liu CC, Yee JL, Tamaki SJ, Marquez DM, Tenvooren I, Wai K, et al. (2023). Dynamic CD8+ T cell responses to cancer immunotherapy in human regional lymph nodes are disrupted in metastatic lymph nodes. Cell 186, 1127–1143.e18. 10.1016/j.cell.2023.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Thommen DS, Koelzer VH, Herzig P, Roller A, Trefny M, Dimeloe S, Kiialainen A, Hanhart J, Schill C, Hess C, et al. (2018). A transcriptionally and functionally distinct PD-1+ CD8+ T cell pool with predictive potential in non-small-cell lung cancer treated with PD-1 blockade. Nat. Med. 24, 994–1004. 10.1038/s41591-018-0057-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Im SJ, Obeng RC, Nasti TH, McManus D, Kamphorst AO, Gunisetty S, Prokhnevska N, Carlisle JW, Yu K, Sica GL, et al. (2023). Characteristics and anatomic location of PD-1+TCF1+ stem-like CD8 T cells in chronic viral infection and cancer. Proc. Natl. Acad. Sci. USA 120, e2221985120. 10.1073/pnas.2221985120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.ElTanbouly MA, and Noelle RJ (2021). Rethinking peripheral T cell tolerance: checkpoints across a T cell’s journey. Nat. Rev. Immunol. 21, 257–267. 10.1038/s41577-020-00454-2. [DOI] [PubMed] [Google Scholar]
- 118.Dikiy S, and Rudensky AY (2023). Principles of regulatory T cell function. Immunity 56, 240–255. 10.1016/j.immuni.2023.01.004. [DOI] [PubMed] [Google Scholar]
- 119.Wing K, Onishi Y, Prieto-Martin P, Yamaguchi T, Miyara M, Fehervari Z, Nomura T, and Sakaguchi S. (2008). CTLA-4 control over Foxp3+ regulatory T cell function. Science 322, 271–275. 10.1126/science.1160062. [DOI] [PubMed] [Google Scholar]
- 120.Zeng H, Yang K, Cloer C, Neale G, Vogel P, and Chi H. (2013). mTORC1 couples immune signals and metabolic programming to establish T(reg)-cell function. Nature 499, 485–490. 10.1038/nature12297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Gocher-Demske AM, Cui J, Szymczak-Workman AL, Vignali KM, Latini JN, Pieklo GP, Kimball JC, Avery L, Cipolla EM, Huckestein BR, et al. (2023). IFNgamma-induction of TH1-like regulatory T cells controls antiviral responses. Nat. Immunol. 24, 841–854. 10.1038/s41590-023-01453-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Koch MA, Tucker-Heard G, Perdue NR, Killebrew JR, Urdahl KB, and Campbell DJ (2009). The transcription factor T-bet controls regulatory T cell homeostasis and function during type 1 inflammation. Nat. Immunol. 10, 595–602. 10.1038/ni.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Levine AG, Mendoza A, Hemmers S, Moltedo B, Niec RE, Schizas M, Hoyos BE, Putintseva EV, Chaudhry A, Dikiy S, et al. (2017). Stability and function of regulatory T cells expressing the transcription factor T-bet. Nature 546, 421–425. 10.1038/nature22360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Chung Y, Tanaka S, Chu F, Nurieva RI, Martinez GJ, Rawal S, Wang YH, Lim H, Reynolds JM, Zhou XH, et al. (2011). Follicular regulatory T cells expressing Foxp3 and Bcl-6 suppress germinal center reactions. Nat. Med. 17, 983–988. 10.1038/nm.2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Linterman MA, Pierson W, Lee SK, Kallies A, Kawamoto S, Rayner TF, Srivastava M, Divekar DP, Beaton L, Hogan JJ, et al. (2011). Foxp3+ follicular regulatory T cells control the germinal center response. Nat. Med. 17, 975–982. 10.1038/nm.2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Tay C, Tanaka A, and Sakaguchi S. (2023). Tumor-infiltrating regulatory T cells as targets of cancer immunotherapy. Cancer Cell 41, 450–465. 10.1016/j.ccell.2023.02.014. [DOI] [PubMed] [Google Scholar]
- 127.Saito T, Nishikawa H, Wada H, Nagano Y, Sugiyama D, Atarashi K, Maeda Y, Hamaguchi M, Ohkura N, Sato E, et al. (2016). Two FOXP3(+)CD4(+) T cell subpopulations distinctly control the prognosis of colorectal cancers. Nat. Med. 22, 679–684. 10.1038/nm.4086. [DOI] [PubMed] [Google Scholar]
- 128.Moreno Ayala MA, Campbell TF, Zhang C, Dahan N, Bockman A, Prakash V, Feng L, Sher T, and DuPage M. (2023). CXCR3 expression in regulatory T cells drives interactions with type I dendritic cells in tumors to restrict CD8+ T cell antitumor immunity. Immunity 56, 1613–1630.e5. 10.1016/j.immuni.2023.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Overacre-Delgoffe AE, Chikina M, Dadey RE, Yano H, Brunazzi EA, Shayan G, Horne W, Moskovitz JM, Kolls JK, Sander C, et al. (2017). Interferon-gamma Drives Treg Fragility to Promote Anti-tumor Immunity. Cell 169, 1130–1141.e11. 10.1016/j.cell.2017.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Watson MJ, Vignali PDA, Mullett SJ, Overacre-Delgoffe AE, Peralta RM, Grebinoski S, Menk AV, Rittenhouse NL, DePeaux K, Whetstone RD, et al. (2021). Metabolic support of tumour-infiltrating regulatory T cells by lactic acid. Nature 591, 645–651. 10.1038/s41586-020-03045-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wang H, Franco F, Tsui YC, Xie X, Trefny MP, Zappasodi R, Mohmood SR, Fernández-Garćıa J, Tsai CH, Schulze I, et al. (2020). CD36-mediated metabolic adaptation supports regulatory T cell survival and function in tumors. Nat. Immunol. 21, 298–308. 10.1038/s41590-019-0589-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lim SA, Wei J, Nguyen TM, Shi H, Su W, Palacios G, Dhungana Y, Chapman NM, Long L, Saravia J, et al. (2021). Lipid signalling enforces functional specialization of Treg cells in tumours. Nature 591, 306–311. 10.1038/s41586-021-03235-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zappasodi R, Serganova I, Cohen IJ, Maeda M, Shindo M, Senbabaoglu Y, Watson MJ, Leftin A, Maniyar R, Verma S, et al. (2021). CTLA-4 blockade drives loss of Treg stability in glycolysis-low tumours. Nature 591, 652–658. 10.1038/s41586-021-03326-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Groux H, O’Garra A, Bigler M, Rouleau M, Antonenko S, de Vries JE, and Roncarolo MG (1997). A CD4+ T-cell subset inhibits antigen-specific T-cell responses and prevents colitis. Nature 389, 737–742. 10.1038/39614. [DOI] [PubMed] [Google Scholar]
- 135.Li J, Zaslavsky M, Su Y, Guo J, Sikora MJ, van Unen V, Christophersen A, Chiou SH, Chen L, Li J, et al. (2022). KIR+CD8+ T cells suppress pathogenic T cells and are active in autoimmune diseases and COVID-19. Science 376, eabi9591. 10.1126/science.abi9591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Mizoguchi A, Mizoguchi E, Smith RN, Preffer FI, and Bhan AK (1997). Suppressive role of B cells in chronic colitis of T cell receptor alpha mutant mice. J. Exp. Med. 186, 1749–1756. 10.1084/jem.186.10.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Masopust D, and Soerens AG (2019). Tissue-Resident T Cells and Other Resident Leukocytes. Annu. Rev. Immunol. 37, 521–546. 10.1146/annurev-immunol-042617-053214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Pruner KB, and Pepper M. (2021). Local memory CD4 T cell niches in respiratory viral infection. J. Exp. Med. 218, e20201733. 10.1084/jem.20201733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Soudja SM, Chandrabos C, Yakob E, Veenstra M, Palliser D, and Lauvau G. (2014). Memory-T-cell-derived interferon-γ instructs potent innate cell activation for protective immunity. Immunity 40, 974–988. 10.1016/j.immuni.2014.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Milner JJ, Toma C, He Z, Kurd NS, Nguyen QP, McDonald B, Quezada L, Widjaja CE, Witherden DA, Crowl JT, et al. (2020). Heterogenous Populations of Tissue-Resident CD8+ T Cells Are Generated in Response to Infection and Malignancy. Immunity 52, 808–824.e7. 10.1016/j.immuni.2020.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Szabo PA, Miron M, and Farber DL (2019). Location, location, location: tissue resident memory T cells in mice and humans. Sci. Immunol. 4, eaas9673. 10.1126/sciimmunol.aas9673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Allie SR, Bradley JE, Mudunuru U, Schultz MD, Graf BA, Lund FE, and Randall TD (2019). The establishment of resident memory B cells in the lung requires local antigen encounter. Nat. Immunol. 20, 97–108. 10.1038/s41590-018-0260-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.MacLean AJ, Richmond N, Koneva L, Attar M, Medina CAP, Thornton EE, Gomes AC, El-Turabi A, Bachmann MF, Rijal P, et al. (2022). Secondary influenza challenge triggers resident memory B cell migration and rapid relocation to boost antibody secretion at infected sites. Immunity 55, 718–733.e8. 10.1016/j.immuni.2022.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Pallett LJ, Davies J, Colbeck EJ, Robertson F, Hansi N, Easom NJW, Burton AR, Stegmann KA, Schurich A, Swadling L, et al. (2017). IL-2high tissue-resident T cells in the human liver: Sentinels for hepatotropic infection. J. Exp. Med. 214, 1567–1580. 10.1084/jem.20162115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Buggert M, Nguyen S, Salgado-Montes de Oca G, Bengsch B, Darko S, Ransier A, Roberts ER, Del Alcazar D, Brody IB, Vella LA, et al. (2018). Identification and characterization of HIV-specific resident memory CD8+ T cells in human lymphoid tissue. Sci. Immunol. 3, eaar4526. 10.1126/sciimmunol.aar4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Jozwik A, Habibi MS, Paras A, Zhu J, Guvenel A, Dhariwal J, Almond M, Wong EHC, Sykes A, Maybeno M, et al. (2015). RSV-specific airway resident memory CD8+ T cells and differential disease severity after experimental human infection. Nat. Commun. 6, 10224. 10.1038/ncomms10224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Heeg M, and Goldrath AW (2023). Insights into phenotypic and functional CD8+ TRM heterogeneity. Immunol. Rev. 316, 8–22. 10.1111/imr.13218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Milner JJ, Toma C, Yu B, Zhang K, Omilusik K, Phan AT, Wang D, Getzler AJ, Nguyen T, Crotty S, et al. (2017). Runx3 programs CD8+ T cell residency in non-lymphoid tissues and tumours. Nature 552, 253–257. 10.1038/nature24993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Park SL, Christo SN, Wells AC, Gandolfo LC, Zaid A, Alexandre YO, Burn TN, Schröder J, Collins N, Han SJ, et al. (2023). Divergent molecular networks program functionally distinct CD8+ skin-resident memory T cells. Science 382, 1073–1079. 10.1126/science.adi8885. [DOI] [PubMed] [Google Scholar]
- 150.Jaiswal A, Verma A, Dannenfelser R, Melssen M, Tirosh I, Izar B, Kim TG, Nirschl CJ, Devi KSP, Olson WC Jr., et al. (2022). An activation to memory differentiation trajectory of tumor-infiltrating lymphocytes informs metastatic melanoma outcomes. Cancer Cell 40, 524–544.e5. 10.1016/j.ccell.2022.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Virassamy B, Caramia F, Savas P, Sant S, Wang J, Christo SN, Byrne A, Clarke K, Brown E, Teo ZL, et al. (2023). Intratumoral CD8+ T cells with a tissue-resident memory phenotype mediate local immunity and immune checkpoint responses in breast cancer. Cancer Cell 41, 585–601.e8. 10.1016/j.ccell.2023.01.004. [DOI] [PubMed] [Google Scholar]
- 152.Beura LK, Fares-Frederickson NJ, Steinert EM, Scott MC, Thompson EA, Fraser KA, Schenkel JM, Vezys V, and Masopust D. (2019). CD4+ resident memory T cells dominate immunosurveillance and orchestrate local recall responses. J. Exp. Med. 216, 1214–1229. 10.1084/jem.20181365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Hondowicz BD, Kim KS, Ruterbusch MJ, Keitany GJ, and Pepper M. (2018). IL-2 is required for the generation of viral-specific CD4+ Th1 tissue-resident memory cells and B cells are essential for maintenance in the lung. Eur. J. Immunol. 48, 80–86. 10.1002/eji.201746928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Iijima N, and Iwasaki A. (2014). T cell memory. A local macrophage chemokine network sustains protective tissue-resident memory CD4 T cells. Science 346, 93–98. 10.1126/science.1257530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Teijaro JR, Turner D, Pham Q, Wherry EJ, Lefranç ois L, and Farber DL (2011). Cutting edge: Tissue-retentive lung memory CD4 T cells mediate optimal protection to respiratory virus infection. J. Immunol. 187, 5510–5514. 10.4049/jimmunol.1102243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Smith NM, Wasserman GA, Coleman FT, Hilliard KL, Yamamoto K, Lipsitz E, Malley R, Dooms H, Jones MR, Quinton LJ, and Mizgerd JP (2018). Regionally compartmentalized resident memory T cells mediate naturally acquired protection against pneumococcal pneumonia. Mucosal Immunol. 11, 220–235. 10.1038/mi.2017.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Glennie ND, Yeramilli VA, Beiting DP, Volk SW, Weaver CT, and Scott P. (2015). Skin-resident memory CD4+ T cells enhance protection against Leishmania major infection. J. Exp. Med. 212, 1405–1414. 10.1084/jem.20142101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Hondowicz BD, An D, Schenkel JM, Kim KS, Steach HR, Krishnamurty AT, Keitany GJ, Garza EN, Fraser KA, Moon JJ, et al. (2016). Interleukin-2-Dependent Allergen-Specific Tissue-Resident Memory Cells Drive Asthma. Immunity 44, 155–166. 10.1016/j.immuni.2015.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Zhao J, Zhao J, Mangalam AK, Channappanavar R, Fett C, Meyerholz DK, Agnihothram S, Baric RS, David CS, and Perlman S. (2016). Airway Memory CD4(+) T Cells Mediate Protective Immunity against Emerging Respiratory Coronaviruses. Immunity 44, 1379–1391. 10.1016/j.immuni.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Son YM, Cheon IS, Wu Y, Li C, Wang Z, Gao X, Chen Y, Takahashi Y, Fu YX, Dent AL, et al. (2021). Tissue-resident CD4+ T helper cells assist the development of protective respiratory B and CD8+ T cell subclinical immune activation and a role for CCR5 in Treg function. Sci. memory responses. Sci. Immunol. 6, eabb6852. 10.1126/sciimmunol.abb6852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Swarnalekha N, Schreiner D, Litzler LC, Iftikhar S, Kirchmeier D, Künzli M, Son YM, Sun J, Moreira EA, and King CG (2021). T resident helper cells promote humoral responses in the lung. Sci. Immunol. 6, eabb6808. 10.1126/sciimmunol.abb6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Rüterbusch MJ, Hondowicz BD, Takehara KK, Pruner KB, Griffith TS, and Pepper M. (2023). Allergen exposure functionally alters influenza-specific CD4+ Th1 memory cells in the lung. J. Exp. Med. 220, e20230112. 10.1084/jem.20230112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Onodera T, Takahashi Y, Yokoi Y, Ato M, Kodama Y, Hachimura S, Kurosaki T, and Kobayashi K. (2012). Memory B cells in the lung participate in protective humoral immune responses to pulmonary influenza virus reinfection. Proc. Natl. Acad. Sci. USA 109, 2485–2490. 10.1073/pnas.1115369109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Joo HM, He Y, and Sangster MY (2008). Broad dispersion and lung localization of virus-specific memory B cells induced by influenza pneumonia. Proc. Natl. Acad. Sci. USA 105, 3485–3490. 10.1073/pnas.0800003105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Adachi Y, Onodera T, Yamada Y, Daio R, Tsuiji M, Inoue T, Kobayashi K, Kurosaki T, Ato M, and Takahashi Y. (2015). Distinct germinal center selection at local sites shapes memory B cell response to viral escape. J. Exp. Med. 212, 1709–1723. 10.1084/jem.20142284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Weisel NM, Joachim SM, Smita S, Callahan D, Elsner RA, Conter LJ, Chikina M, Farber DL, Weisel FJ, and Shlomchik MJ (2022). Surface phenotypes of naive and memory B cells in mouse and human tissues. Nat. Immunol. 23, 135–145. 10.1038/s41590-021-01078-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Feuerer M, Herrero L, Cipolletta D, Naaz A, Wong J, Nayer A, Lee J, Goldfine AB, Benoist C, Shoelson S, and Mathis D. (2009). Lean, but not obese, fat is enriched for a unique population of regulatory T cells that affect metabolic parameters. Nat. Med. 15, 930–939. 10.1038/nm.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Cipolletta D, Feuerer M, Li A, Kamei N, Lee J, Shoelson SE, Benoist C, and Mathis D. (2012). PPAR-gamma is a major driver of the accumulation and phenotype of adipose tissue Treg cells. Nature 486, 549–553. 10.1038/nature11132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Vasanthakumar A, Moro K, Xin A, Liao Y, Gloury R, Kawamoto S, Fagarasan S, Mielke LA, Afshar-Sterle S, Masters SL, et al. (2015). The transcriptional regulators IRF4, BATF and IL-33 orchestrate development and maintenance of adipose tissue-resident regulatory T cells. Nat. Immunol. 16, 276–285. 10.1038/ni.3085. [DOI] [PubMed] [Google Scholar]
- 170.Li C, DiSpirito JR, Zemmour D, Spallanzani RG, Kuswanto W, Benoist C, and Mathis D. (2018). TCR Transgenic Mice Reveal Stepwise, Multi-site Acquisition of the Distinctive Fat-Treg Phenotype. Cell 174, 285–299.e12. 10.1016/j.cell.2018.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Miragaia RJ, Gomes T, Chomka A, Jardine L, Riedel A, Hegazy AN, Whibley N, Tucci A, Chen X, Lindeman I, et al. (2019). Single-Cell Transcriptomics of Regulatory T Cells Reveals Trajectories of Tissue Adaptation. Immunity 50, 493–504.e7. 10.1016/j.immuni.2019.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.DiSpirito JR, Zemmour D, Ramanan D, Cho J, Zilionis R, Klein AM, Benoist C, and Mathis D. (2018). Molecular diversification of regulatory T cells in nonlymphoid tissues. Sci. Immunol. 3, eaat5861. 10.1126/sciimmunol.aat5861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Ali N, Zirak B, Rodriguez RS, Pauli ML, Truong HA, Lai K, Ahn R, Corbin K, Lowe MM, Scharschmidt TC, et al. (2017). Regulatory T Cells in Skin Facilitate Epithelial Stem Cell Differentiation. Cell 169, 1119–1129.e11. 10.1016/j.cell.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Gellatly KJ, Strassner JP, Essien K, Refat MA, Murphy RL, Coffin-Schmitt A, Pandya AG, Tovar-Garza A, Frisoli ML, Fan X, et al. (2021). scRNA-seq of human vitiligo reveals complex networks of Transl. Med. 13, eabd8995. 10.1126/scitranslmed.abd8995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Burzyn D, Kuswanto W, Kolodin D, Shadrach JL, Cerletti M, Jang Y, Sefik E, Tan TG, Wagers AJ, Benoist C, and Mathis D. (2013). A special population of regulatory T cells potentiates muscle repair. Cell 155, 1282–1295. 10.1016/j.cell.2013.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Arpaia N, Green JA, Moltedo B, Arvey A, Hemmers S, Yuan S, Treuting PM, and Rudensky AY (2015). A Distinct Function of Regulatory T Cells in Tissue Protection. Cell 162, 1078–1089. 10.1016/j.cell.2015.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Kaiser KA, Loffredo LF, Santos-Alexis KL, Ringham OR, and Arpaia N. (2023). Regulation of the alveolar regenerative niche by amphiregulin-producing regulatory T cells. J. Exp. Med. 220, e20221462. 10.1084/jem.20221462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Ito M, Komai K, Mise-Omata S, Iizuka-Koga M, Noguchi Y, Kondo T, Sakai R, Matsuo K, Nakayama T, Yoshie O, et al. (2019). Brain regulatory T cells suppress astrogliosis and potentiate neurological recovery. Nature 565, 246–250. 10.1038/s41586-018-0824-5. [DOI] [PubMed] [Google Scholar]
- 179.Brown CC, and Rudensky AY (2023). Spatiotemporal regulation of peripheral T cell tolerance. Science 380, 472–478. 10.1126/science.adg6425. [DOI] [PubMed] [Google Scholar]
- 180.Akagbosu B, Tayyebi Z, Shibu G, Paucar Iza YA, Deep D, Parisotto YF, Fisher L, Pasolli HA, Thevin V, Elmentaite R, et al. (2022). Novel antigen-presenting cell imparts Treg-dependent tolerance to gut microbiota. Nature 610, 752–760. 10.1038/s41586-022-05309-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Kedmi R, Najar TA, Mesa KR, Grayson A, Kroehling L, Hao Y, Hao S, Pokrovskii M, Xu M, Talbot J, et al. (2022). A RORγt+ cell instructs gut microbiota-specific Treg cell differentiation. Nature 610, 737–743. 10.1038/s41586-022-05089-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Lyu M, Suzuki H, Kang L, Gaspal F, Zhou W, Goc J, Zhou L, Zhou J, Zhang W, et al. ; JRI Live Cell Bank (2022). ILC3s select microbiota-specific regulatory T cells to establish tolerance in the gut. Nature 610, 744–751. 10.1038/s41586-022-05141-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Meizlish ML, Franklin RA, Zhou X, and Medzhitov R. (2021). Tissue Homeostasis and Inflammation. Annu. Rev. Immunol. 39, 557–581. 10.1146/annurev-immunol-061020-053734. [DOI] [PubMed] [Google Scholar]
- 184.Rankin LC, and Artis D. (2018). Beyond Host Defense: Emerging Functions of the Immune System in Regulating Complex Tissue Physiology. Cell 173, 554–567. 10.1016/j.cell.2018.03.013. [DOI] [PubMed] [Google Scholar]
- 185.Kioussis D, and Pachnis V. (2009). Immune and nervous systems: more than just a superficial similarity? Immunity 31, 705–710. 10.1016/j.immuni.2009.09.009. [DOI] [PubMed] [Google Scholar]
- 186.Enamorado M, Kulalert W, Han SJ, Rao I, Delaleu J, Link VM, Yong D, Smelkinson M, Gil L, Nakajima S, et al. (2023). Immunity to the microbiota promotes sensory neuron regeneration. Cell 186, 607–620.e17. 10.1016/j.cell.2022.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Florsheim EB, Bachtel ND, Cullen JL, Lima BGC, Godazgar M, Carvalho F, Chatain CP, Zimmer MR, Zhang C, Gautier G, et al. (2023). Immune sensing of food allergens promotes avoidance behaviour. Nature 620, 643–650. 10.1038/s41586-023-06362-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Talbot S, Abdulnour RE, Burkett PR, Lee S, Cronin SJ, Pascal MA, Laedermann C, Foster SL, Tran JV, Lai N, et al. (2015). Silencing Nociceptor Neurons Reduces Allergic Airway Inflammation. Neuron 87, 341–354. 10.1016/j.neuron.2015.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Wang L, Cheng M, Wang Y, Chen J, Xie F, Huang LH, and Zhan C. (2024). Fasting-activated ventrolateral medulla neurons regulate T cell homing and suppress autoimmune disease in mice. Nat. Neurosci. 27, 462–470. 10.1038/s41593-023-01543-w. [DOI] [PubMed] [Google Scholar]
- 190.Collins N, Han SJ, Enamorado M, Link VM, Huang B, Moseman EA, Kishton RJ, Shannon JP, Dixit D, Schwab SR, et al. (2019). The Bone Marrow Protects and Optimizes Immunological Memory during Dietary Restriction. Cell 178, 1088–1101.e15. 10.1016/j.cell.2019.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Globig AM, Zhao S, Roginsky J, Maltez VI, Guiza J, Avina-Ochoa N, Heeg M, Araujo Hoffmann F, Chaudhary O, Wang J, et al. (2023). The β1-adrenergic receptor links sympathetic nerves to T cell exhaustion. Nature 622, 383–392. 10.1038/s41586-023-06568-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Stinchcombe JC, Asano Y, Kaufman CJG, Böhlig K, Peddie CJ, Collinson LM, Nadler A, and Griffiths GM (2023). Ectocytosis renders T cell receptor signaling self-limiting at the immune synapse. Science 380, 818–823. 10.1126/science.abp8933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Liu B, Chen W, Evavold BD, and Zhu C. (2014). Accumulation of dynamic catch bonds between TCR and agonist peptide-MHC triggers T cell signaling. Cell 157, 357–368. 10.1016/j.cell.2014.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Man K, Miasari M, Shi W, Xin A, Henstridge DC, Preston S, Pellegrini M, Belz GT, Smyth GK, Febbraio MA, et al. (2013). The transcription factor IRF4 is essential for TCR affinity-mediated metabolic programming and clonal expansion of T cells. Nat. Immunol. 14, 1155–1165. 10.1038/ni.2710. [DOI] [PubMed] [Google Scholar]
- 195.Yao S, Buzo BF, Pham D, Jiang L, Taparowsky EJ, Kaplan MH, and Sun J. (2013). Interferon regulatory factor 4 sustains CD8(+) T cell expansion and effector differentiation. Immunity 39, 833–845. 10.1016/j.immuni.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Krishnamoorthy V, Kannanganat S, Maienschein-Cline M, Cook SL, Chen J, Bahroos N, Sievert E, Corse E, Chong A, and Sciammas R. (2017). The IRF4 Gene Regulatory Module Functions as a Read-Write Integrator to Dynamically Coordinate T Helper Cell Fate. Immunity 47, 481–497.e7. 10.1016/j.immuni.2017.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Ochiai K, Maienschein-Cline M, Simonetti G, Chen J, Rosenthal R, Brink R, Chong AS, Klein U, Dinner AR, Singh H, and Sciammas R. (2013). Transcriptional regulation of germinal center B and plasma cell fates by dynamical control of IRF4. Immunity 38, 918–929. 10.1016/j.immuni.2013.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Klein Geltink RI, O’Sullivan D, Corrado M, Bremser A, Buck MD, Buescher JM, Firat E, Zhu X, Niedermann G, Caputa G, et al. (2017). Mitochondrial Priming by CD28. Cell 171, 385–397.e11. 10.1016/j.cell.2017.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Yang K, Shrestha S, Zeng H, Karmaus PW, Neale G, Vogel P, Guertin DA, Lamb RF, and Chi H. (2013). T cell exit from quiescence and differentiation into Th2 cells depend on Raptor-mTORC1-mediated metabolic reprogramming. Immunity 39, 1043–1056. 10.1016/j.immuni.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Gigoux M, Shang J, Pak Y, Xu M, Choe J, Mak TW, and Suh WK (2009). Inducible costimulator promotes helper T-cell differentiation through phosphoinositide 3-kinase. Proc. Natl. Acad. Sci. USA 106, 20371–20376. 10.1073/pnas.0911573106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Kamphorst AO, Wieland A, Nasti T, Yang S, Zhang R, Barber DL, Konieczny BT, Daugherty CZ, Koenig L, Yu K, et al. (2017). Rescue of exhausted CD8 T cells by PD-1-targeted therapies is CD28-dependent. Science 355, 1423–1427. 10.1126/science.aaf0683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Hashimoto M, Araki K, Cardenas MA, Li P, Jadhav RR, Kissick HT, Hudson WH, McGuire DJ, Obeng RC, Wieland A, et al. (2022). PD-1 combination therapy with IL-2 modifies CD8+ T cell exhaustion program. Nature 610, 173–181. 10.1038/s41586-022-05257-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Codarri Deak L, Nicolini V, Hashimoto M, Karagianni M, Schwalie PC, Lauener L, Varypataki EM, Richard M, Bommer E, Sam J, et al. (2022). PD-1-cis IL-2R agonism yields better effectors from stem--like CD8+ T cells. Nature 610, 161–172. 10.1038/s41586-022-05192-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Lee JY, Hall JA, Kroehling L, Wu L, Najar T, Nguyen HH, Lin WY, Yeung ST, Silva HM, Li D, et al. (2020). Serum Amyloid A Proteins Induce Pathogenic Th17 Cells and Promote Inflammatory Disease. Cell 180, 79–91.e16. 10.1016/j.cell.2019.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Long L, Wei J, Lim SA, Raynor JL, Shi H, Connelly JP, Wang H, Guy C, Xie B, Chapman NM, et al. (2021). CRISPR screens unveil signal hubs for nutrient licensing of T cell immunity. Nature 600, 308–313. 10.1038/s41586-021-04109-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Raynor JL, and Chi H. (2024). Nutrients: Signal 4 in T cell immunity. J. Exp. Med. 221, e20221839. 10.1084/jem.20221839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Reinfeld BI, Madden MZ, Wolf MM, Chytil A, Bader JE, Patterson AR, Sugiura A, Cohen AS, Ali A, Do BT, et al. (2021). Cell-programmed nutrient partitioning in the tumour microenvironment. Nature 593, 282–288. 10.1038/s41586-021-03442-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Ho PC, Bihuniak JD, Macintyre AN, Staron M, Liu X, Amezquita R, Tsui YC, Cui G, Micevic G, Perales JC, et al. (2015). Phosphoenolpyruvate Is a Metabolic Checkpoint of Anti-tumor T Cell Responses. Cell 162, 1217–1228. 10.1016/j.cell.2015.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Chang CH, Qiu J, O’Sullivan D, Buck MD, Noguchi T, Curtis JD, Chen Q, Gindin M, Gubin MM, van der Windt GJ, et al. (2015). Metabolic Competition in the Tumor Microenvironment Is a Driver of Cancer Progression. Cell 162, 1229–1241. 10.1016/j.cell.2015.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Wang R, Dillon CP, Shi LZ, Milasta S, Carter R, Finkelstein D, McCormick LL, Fitzgerald P, Chi H, Munger J, and Green DR (2011). The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity 35, 871–882. 10.1016/j.immuni.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Johnson MO, Wolf MM, Madden MZ, Andrejeva G, Sugiura A, Contreras DC, Maseda D, Liberti MV, Paz K, Kishton RJ, et al. (2018). Distinct Regulation of Th17 and Th1 Cell Differentiation by Glutaminase-Dependent Metabolism. Cell 175, 1780–1795.e19. 10.1016/j.cell.2018.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Leone RD, Zhao L, Englert JM, Sun IM, Oh MH, Sun IH, Arwood ML, Bettencourt IA, Patel CH, Wen J, et al. (2019). Glutamine blockade induces divergent metabolic programs to overcome tumor immune evasion. Science 366, 1013–1021. 10.1126/science.aav2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Guo C, You Z, Shi H, Sun Y, Du X, Palacios G, Guy C, Yuan S, Chapman NM, Lim SA, et al. (2023). SLC38A2 and glutamine signalling in cDC1s dictate anti-tumour immunity. Nature 620, 200–208. 10.1038/s41586-023-06299-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Geiger R, Rieckmann JC, Wolf T, Basso C, Feng Y, Fuhrer T, Kogadeeva M, Picotti P, Meissner F, Mann M, et al. (2016). L-Arginine Modulates T Cell Metabolism and Enhances Survival and Anti-tumor Activity. Cell 167, 829–842.e13. 10.1016/j.cell.2016.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Bian Y, Li W, Kremer DM, Sajjakulnukit P, Li S, Crespo J, Nwosu ZC, Zhang L, Czerwonka A, Pawłowska A, et al. (2020). Cancer SLC43A2 alters T cell methionine metabolism and histone methylation. Nature 585, 277–282. 10.1038/s41586-020-2682-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Shi H, Chapman NM, Wen J, Guy C, Long L, Dhungana Y, Rankin S, Pelletier S, Vogel P, Wang H, et al. (2019). Amino Acids License Kinase mTORC1 Activity and Treg Cell Function via Small G Proteins Rag and Rheb. Immunity 51, 1012–1027.e7. 10.1016/j.immuni.2019.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Brand A, Singer K, Koehl GE, Kolitzus M, Schoenhammer G, Thiel A, Matos C, Bruss C, Klobuch S, Peter K, et al. (2016). LDHA-Associated Lactic Acid Production Blunts Tumor Immunosurveillance by T and NK Cells. Cell Metab. 24, 657–671. 10.1016/j.cmet.2016.08.011. [DOI] [PubMed] [Google Scholar]
- 218.Xu S, Chaudhary O, Rodríguez-Morales P, Sun X, Chen D, Zappasodi R, Xu Z, Pinto AFM, Williams A, Schulze I, et al. (2021). Uptake of oxidized lipids by the scavenger receptor CD36 promotes lipid peroxidation and dysfunction in CD8+ T cells in tumors. Immunity 54, 1561–1577.e7. 10.1016/j.immuni.2021.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Wu VH, Yung BS, Faraji F, Saddawi-Konefka R, Wang Z, Wenzel AT, Song MJ, Pagadala MS, Clubb LM, Chiou J, et al. (2023). The GPCR-Gαs-PKA signaling axis promotes T cell dysfunction and cancer immunotherapy failure. Nat. Immunol. 24, 1318–1330. 10.1038/s41590-023-01529-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Fan H, Xia S, Xiang J, Li Y, Ross MO, Lim SA, Yang F, Tu J, Xie L, Dougherty U, et al. (2023). Trans-vaccenic acid reprograms CD8+ T cells and anti-tumour immunity. Nature 623, 1034–1043. 10.1038/s41586-023-06749-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Macián F, García-Cózar F, Im SH, Horton HF, Byrne MC, and Rao A. (2002). Transcriptional mechanisms underlying lymphocyte tolerance. Cell 109, 719–731. 10.1016/s0092-8674(02)00767-5. [DOI] [PubMed] [Google Scholar]
- 222.Naramura M, Jang IK, Kole H, Huang F, Haines D, and Gu H. (2002). c-Cbl and Cbl-b regulate T cell responsiveness by promoting ligand-induced TCR down-modulation. Nat. Immunol. 3, 1192–1199. 10.1038/ni855. [DOI] [PubMed] [Google Scholar]
- 223.Uehata T, Iwasaki H, Vandenbon A, Matsushita K, Hernandez-Cuellar E, Kuniyoshi K, Satoh T, Mino T, Suzuki Y, Standley DM, et al. (2013). Malt1-induced cleavage of regnase-1 in CD4(+) helper T cells regulates immune activation. Cell 153, 1036–1049. 10.1016/j.cell.2013.04.034. [DOI] [PubMed] [Google Scholar]
- 224.Michalek RD, Gerriets VA, Jacobs SR, Macintyre AN, MacIver NJ, Mason EF, Sullivan SA, Nichols AG, and Rathmell JC (2011). Cutting edge: distinct glycolytic and lipid oxidative metabolic programs are essential for effector and regulatory CD4+ T cell subsets. J. Immunol. 186, 3299–3303. 10.4049/jimmunol.1003613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Pearce EL, Walsh MC, Cejas PJ, Harms GM, Shen H, Wang LS, Jones RG, and Choi Y. (2009). Enhancing CD8 T-cell memory by modulating fatty acid metabolism. Nature 460, 103–107. 10.1038/nature08097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Scharping NE, Rivadeneira DB, Menk AV, Vignali PDA, Ford BR, Rittenhouse NL, Peralta R, Wang Y, Wang Y, DePeaux K, et al. (2021). Mitochondrial stress induced by continuous stimulation under hypoxia rapidly drives T cell exhaustion. Nat. Immunol. 22, 205–215. 10.1038/s41590-020-00834-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Yu YR, Imrichova H, Wang H, Chao T, Xiao Z, Gao M, Rincon-Restrepo M, Franco F, Genolet R, Cheng WC, et al. (2020). Disturbed mitochondrial dynamics in CD8+ TILs reinforce T cell exhaustion. Nat. Immunol. 21, 1540–1551. 10.1038/s41590-020-0793-3. [DOI] [PubMed] [Google Scholar]
- 228.Vardhana SA, Hwee MA, Berisa M, Wells DK, Yost KE, King B, Smith M, Herrera PS, Chang HY, Satpathy AT, et al. (2020). Impaired mitochondrial oxidative phosphorylation limits the self-renewal of T cells exposed to persistent antigen. Nat. Immunol. 21, 1022–1033. 10.1038/s41590-020-0725-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Jaccard A, Wyss T, Maldonado-Pérez N, Rath JA, Bevilacqua A, Peng JJ, Lepez A, Von Gunten C, Franco F, Kao KC, et al. (2023). Reductive carboxylation epigenetically instructs T cell differentiation. Nature 621, 849–856. 10.1038/s41586-023-06546-y. [DOI] [PubMed] [Google Scholar]
- 230.Notarangelo G, Spinelli JB, Perez EM, Baker GJ, Kurmi K, Elia I, Stopka SA, Baquer G, Lin JR, Golby AJ, et al. (2022). Oncometabolite d-2HG alters T cell metabolism to impair CD8+ T cell function. Science 377, 1519–1529. 10.1126/science.abj5104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Huang H, Zhou P, Wei J, Long L, Shi H, Dhungana Y, Chapman NM, Fu G, Saravia J, Raynor JL, et al. (2021). In vivo CRISPR screening reveals nutrient signaling processes underpinning CD8+ T cell fate decisions. Cell 184, 1245–1261.e21. 10.1016/j.cell.2021.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Fu G, Guy CS, Chapman NM, Palacios G, Wei J, Zhou P, Long L, Wang YD, Qian C, Dhungana Y, et al. (2021). Metabolic control of TFH cells and humoral immunity by phosphatidylethanolamine. Nature 595, 724–729. 10.1038/s41586-021-03692-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Sen DR, Kaminski J, Barnitz RA, Kurachi M, Gerdemann U, Yates KB, Tsao HW, Godec J, LaFleur MW, Brown FD, et al. (2016). The epigenetic landscape of T cell exhaustion. Science 354, 1165–1169. 10.1126/science.aae0491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Pauken KE, Sammons MA, Odorizzi PM, Manne S, Godec J, Khan O, Drake AM, Chen Z, Sen DR, Kurachi M, et al. (2016). Epigenetic stability of exhausted T cells limits durability of reinvigoration by PD-1 blockade. Science 354, 1160–1165. 10.1126/science.aaf2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Scott-Browne JP, López-Moyado IF, Trifari S, Wong V, Chavez L, Rao A, and Pereira RM (2016). Dynamic Changes in Chromatin Accessibility Occur in CD8+ T Cells Responding to Viral. Immunity 45, 1327–1340. 10.1016/j.immuni.2016.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Philip M, Fairchild L, Sun L, Horste EL, Camara S, Shakiba M, Scott AC, Viale A, Lauer P, Merghoub T, et al. (2017). Chromatin states define tumour-specific T cell dysfunction and reprogramming. Nature 545, 452–456. 10.1038/nature22367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Mognol GP, Spreafico R, Wong V, Scott-Browne JP, Togher S, Hoffmann A, Hogan PG, Rao A, and Trifari S. (2017). Exhaustion-associated regulatory regions in CD8+ tumor-infiltrating T cells. Proc. Natl. Acad. Sci. USA 114, E2776–E2785. 10.1073/pnas.1620498114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Yates KB, Tonnerre P, Martin GE, Gerdemann U, Al Abosy R, Comstock DE, Weiss SA, Wolski D, Tully DC, Chung RT, et al. (2021). Epigenetic scars of CD8+ T cell exhaustion persist after cure of chronic infection in humans. Nat. Immunol. 22, 1020–1029. 10.1038/s41590-021-00979-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Wei G, Wei L, Zhu J, Zang C, Hu-Li J, Yao Z, Cui K, Kanno Y, Roh TY, Watford WT, et al. (2009). Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity 30, 155–167. 10.1016/j.immuni.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Ghoneim HE, Fan Y, Moustaki A, Abdelsamed HA, Dash P, Dogra P, Carter R, Awad W, Neale G, Thomas PG, and Young-blood B. (2017). De Novo Epigenetic Programs Inhibit PD-1 Blockade-Mediated T Cell Rejuvenation. Cell 170, 142–157.e19. 10.1016/j.cell.2017.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Guo A, Huang H, Zhu Z, Chen MJ, Shi H, Yuan S, Sharma P, Connelly JP, Liedmann S, Dhungana Y, et al. (2022). cBAF complex components and MYC cooperate early in CD8+ T cell fate. Nature 607, 135–141. 10.1038/s41586-022-04849-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Belk JA, Yao W, Ly N, Freitas KA, Chen YT, Shi Q, Valencia AM, Shifrut E, Kale N, Yost KE, et al. (2022). Genome-wide CRISPR screens of T cell exhaustion identify chromatin remodeling factors that limit T cell persistence. Cancer Cell 40, 768–786.e7. 10.1016/j.ccell.2022.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Baxter AE, Huang H, Giles JR, Chen Z, Wu JE, Drury S, Dalton K, Park SL, Torres L, Simone BW, et al. (2023). The SWI/SNF chromatin remodeling complexes BAF and PBAF differentially regulate epigenetic transitions in exhausted CD8+ T cells. Immunity 56, 1320–1340.e10. 10.1016/j.immuni.2023.05.008. [DOI] [PubMed] [Google Scholar]
- 244.McDonald B, Chick BY, Ahmed NS, Burns M, Ma S, Casillas E, Chen D, Mann TH, O’Connor C, Hah N, et al. (2023). Canonical BAF complex activity shapes the enhancer landscape that licenses CD8+ T cell effector and memory fates. Immunity 56, 1303–1319.e5. 10.1016/j.immuni.2023.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Loo CS, Gatchalian J, Liang Y, Leblanc M, Xie M, Ho J, Venkatraghavan B, Hargreaves DC, and Zheng Y. (2020). A Genome-wide CRISPR Screen Reveals a Role for the Non-canonical Nucleosome-Remodeling BAF Complex in Foxp3 Expression and Regulatory T Cell Function. Immunity 53, 143–157.e8. 10.1016/j.immuni.2020.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Riedel S. (2005). Edward Jenner and the history of smallpox and vaccination. Proc. (Bayl Univ. Med. Cent) 18, 21–25. 10.1080/08998280.2005.11928028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Fong Y, Huang Y, Benkeser D, Carpp LN, Áñez G, Woo W, McGarry A, Dunkle LM, Cho I, Houchens CR, et al. (2023). Immune correlates analysis of the PREVENT-19 COVID-19 vaccine efficacy clinical trial. Nat. Commun. 14, 331. 10.1038/s41467-022-35768-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Barouch DH (2022). Covid-19 Vaccines - Immunity, Variants, Boosters. N. Engl. J. Med. 387, 1011–1020. 10.1056/NEJMra2206573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Sette A, Sidney J, and Crotty S. (2023). T Cell Responses to SARS-CoV-2. Annu. Rev. Immunol. 41, 343–373. 10.1146/annurev-immunol-101721-061120. [DOI] [PubMed] [Google Scholar]
- 250.Amanat F, Thapa M, Lei T, Ahmed SMS, Adelsberg DC, Carreño JM, Strohmeier S, Schmitz AJ, Zafar S, Zhou JQ, et al. (2021). SARS-CoV-2 mRNA vaccination induces functionally diverse antibodies to NTD, RBD, and S2. Cell 184, 3936–3948.e10. 10.1016/j.cell.2021.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Turner JS, O’Halloran JA, Kalaidina E, Kim W, Schmitz AJ, Zhou JQ, Lei T, Thapa M, Chen RE, Case JB, et al. (2021). SARS-CoV-2 mRNA vaccines induce persistent human germinal centre responses. Nature 596, 109–113. 10.1038/s41586-021-03738-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Mudd PA, Minervina AA, Pogorelyy MV, Turner JS, Kim W, Kalaidina E, Petersen J, Schmitz AJ, Lei T, Haile A, et al. (2022). SARS-CoV-2 mRNA vaccination elicits a robust and persistent T follicular helper cell response in humans. Cell 185, 603–613.e15. 10.1016/j.cell.2021.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Turner JS, Kim W, Kalaidina E, Goss CW, Rauseo AM, Schmitz AJ, Hansen L, Haile A, Klebert MK, Pusic I, et al. (2021). SARS-CoV-2 infection induces long-lived bone marrow plasma cells in humans. Nature 595, 421–425. 10.1038/s41586-021-03647-4. [DOI] [PubMed] [Google Scholar]
- 254.Goel RR, Painter MM, Apostolidis SA, Mathew D, Meng W, Rosenfeld AM, Lundgreen KA, Reynaldi A, Khoury DS, Pattekar A, et al. (2021). mRNA vaccines induce durable immune memory to SARS-CoV-2 and variants of concern. Science 374, abm0829. 10.1126/science.abm0829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Rodda LB, Netland J, Shehata L, Pruner KB, Morawski PA, Thouvenel CD, Takehara KK, Eggenberger J, Hemann EA, Waterman HR, et al. (2021). Functional SARS-CoV-2-Specific Immune Memory Persists after Mild COVID-19. Cell 184, 169–183.e17. 10.1016/j.cell.2020.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Bellusci L, Grubbs G, Zahra FT, Forgacs D, Golding H, Ross TM, and Khurana S. (2022). Antibody affinity and cross-variant neutralization of SARS-CoV-2 Omicron BA.1, BA.2 and BA.3 following third mRNA vaccination. Nat. Commun. 13, 4617. 10.1038/s41467-022-32298-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Bange EM, Han NA, Wileyto P, Kim JY, Gouma S, Robinson J, Greenplate AR, Hwee MA, Porterfield F, Owoyemi O, et al. (2021). CD8+ T cells contribute to survival in patients with COVID-19 and hematologic cancer. Nat. Med. 27, 1280–1289. 10.1038/s41591-021-01386-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Janeway CA Jr. (1989). Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb. Symp. Quant. Biol. 54, 1–13. 10.1101/sqb.1989.054.01.003. [DOI] [PubMed] [Google Scholar]
- 259.Facciolá A, Visalli G, Laganá A, and Di Pietro A. (2022). An Overview of Vaccine Adjuvants: Current Evidence and Future Perspectives. Vaccines (Basel) 10, 819. 10.3390/vaccines10050819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Lavelle EC, and Ward RW (2022). Mucosal vaccines - fortifying the frontiers. Nat. Rev. Immunol. 22, 236–250. 10.1038/s41577-021-00583-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Lin MJ, Svensson-Arvelund J, Lubitz GS, Marabelle A, Melero I, Brown BD, and Brody JD (2022). Cancer vaccines: the next immuno-therapy frontier. Nat. Cancer 3, 911–926. 10.1038/s43018-022-00418-6. [DOI] [PubMed] [Google Scholar]
- 262.Rojas LA, Sethna Z, Soares KC, Olcese C, Pang N, Patterson E, Lihm J, Ceglia N, Guasp P, Chu A, et al. (2023). Personalized RNA neoantigen vaccines stimulate T cells in pancreatic cancer. Nature 618, 144–150. 10.1038/s41586-023-06063-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Lei J, Ploner A, Elfström KM, Wang J, Roth A, Fang F, Sundström K, Dillner J, and Sparén P. (2020). HPV Vaccination and the Risk of Invasive Cervical Cancer. N. Engl. J. Med. 383, 1340–1348. 10.1056/NEJMoa1917338. [DOI] [PubMed] [Google Scholar]
- 264.Irvine DJ, Maus MV, Mooney DJ, and Wong WW (2022). The future of engineered immune cell therapies. Science 378, 853–858. 10.1126/science.abq6990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Rosenberg SA, Packard BS, Aebersold PM, Solomon D, Topalian SL, Toy ST, Simon P, Lotze MT, Yang JC, Seipp CA, et al. (1988). Use of tumor-infiltrating lymphocytes and interleukin-2 in the immunotherapy of patients with metastatic melanoma. A preliminary report. N. Engl. J. Med. 319, 1676–1680. 10.1056/NEJM198812223192527. [DOI] [PubMed] [Google Scholar]
- 266.Porter DL, Levine BL, Kalos M, Bagg A, and June CH (2011). Chimeric antigen receptor-modified T cells in chronic lymphoid leukemia. N. Engl. J. Med. 365, 725–733. 10.1056/NEJMoa1103849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Brentjens RJ, Rivière I, Park JH, Davila ML, Wang X, Stefanski J, Taylor C, Yeh R, Bartido S, Borquez-Ojeda O, et al. (2011). Safety and persistence of adoptively transferred autologous CD19-targeted T cells in patients with relapsed or chemotherapy refractory B-cell leukemias. Blood 118, 4817–4828. 10.1182/blood-2011-04-348540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Kochenderfer JN, Wilson WH, Janik JE, Dudley ME, Stetler-Stevenson M, Feldman SA, Maric I, Raffeld M, Nathan DA, Lanier BJ, et al. (2010). Eradication of B-lineage cells and regression of lymphoma in a patient treated with autologous T cells genetically engineered to recognize CD19. Blood 116, 4099–4102. 10.1182/blood-2010-04-281931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Kloss CC, Condomines M, Cartellieri M, Bachmann M, and Sadelain M. (2013). Combinatorial antigen recognition with balanced signaling promotes selective tumor eradication by engineered T cells. Nat. Bio-technol. 31, 71–75. 10.1038/nbt.2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Tousley AM, Rotiroti MC, Labanieh L, Rysavy LW, Kim WJ, Lareau C, Sotillo E, Weber EW, Rietberg SP, Dalton GN, et al. (2023). Co-opting signalling molecules enables logic-gated control of CAR T cells. Nature 615, 507–516. 10.1038/s41586-023-05778-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Shi H, Doench JG, and Chi H. (2023). CRISPR screens for functional interrogation of immunity. Nat. Rev. Immunol. 23, 363–380. 10.1038/s41577-022-00802-4. [DOI] [PubMed] [Google Scholar]
- 272.Dong MB, Wang G, Chow RD, Ye L, Zhu L, Dai X, Park JJ, Kim HR, Errami Y, Guzman CD, et al. (2019). Systematic Immunotherapy Target Discovery Using Genome-Scale In Vivo CRISPR Screens in CD8 T Cells. Cell 178, 1189–1204.e23. 10.1016/j.cell.2019.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Wei J, Long L, Zheng W, Dhungana Y, Lim SA, Guy C, Wang Y, Wang YD, Qian C, Xu B, et al. (2019). Targeting REGNASE-1 programs long-lived effector T cells for cancer therapy. Nature 576, 471–476. 10.1038/s41586-019-1821-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Carnevale J, Shifrut E, Kale N, Nyberg WA, Blaeschke F, Chen YY, Li Z, Bapat SP, Diolaiti ME, O’Leary P, et al. (2022). RASA2 ablation in T cells boosts antigen sensitivity and long-term function. Nature 609, 174–182. 10.1038/s41586-022-05126-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Larson RC, Kann MC, Bailey SR, Haradhvala NJ, Llopis PM, Bouffard AA, Scarfó I, Leick MB, Grauwet K, Berger TR, et al. (2022). CAR T cell killing requires the IFNgR pathway in solid but not liquid tumours. Nature 604, 563–570. 10.1038/s41586-022-04585-5. [DOI] [PubMed] [Google Scholar]
- 276.Eyquem J, Mansilla-Soto J, Giavridis T, van der Stegen SJ, Hamieh M, Cunanan KM, Odak A, Gönen M, and Sadelain M. (2017). Targeting a CAR to the TRAC locus with CRISPR/Cas9 enhances tumour rejection. Nature 543, 113–117. 10.1038/nature21405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Zhang J, Hu Y, Yang J, Li W, Zhang M, Wang Q, Zhang L, Wei G, Tian Y, Zhao K, et al. (2022). Non-viral, specifically targeted CAR-T cells achieve high safety and efficacy in B-NHL. Nature 609, 369–374. 10.1038/s41586-022-05140-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Hurton LV, Singh H, Najjar AM, Switzer KC, Mi T, Maiti S, Olivares S, Rabinovich B, Huls H, Forget MA, et al. (2016). Tethered IL-15 augments antitumor activity and promotes a stem-cell memory subset in tumor-specific T cells. Proc. Natl. Acad. Sci. USA 113, E7788–E7797. 10.1073/pnas.1610544113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Zhao Y, Chen J, Andreatta M, Feng B, Xie YQ, Wenes M, Wang Y, Gao M, Hu X, Romero P, et al. (2024). IL-10-expressing CAR T cells resist dysfunction and mediate durable clearance of solid tumors and metastases. Published online January 2, 2024. Nat. Biotechnol. 10.1038/s41587-023-02060-8. [DOI] [PubMed] [Google Scholar]
- 280.Kloss CC, Lee J, Zhang A, Chen F, Melenhorst JJ, Lacey SF, Maus MV, Fraietta JA, Zhao Y, and June CH (2018). Dominant-Negative TGF-beta Receptor Enhances PSMA-Targeted Human CAR T Cell Proliferation And Augments Prostate Cancer Eradication. Mol. Ther. 26, 1855–1866. 10.1016/j.ymthe.2018.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Rafiq S, Yeku OO, Jackson HJ, Purdon TJ, van Leeuwen DG, Drakes DJ, Song M, Miele MM, Li Z, Wang P, et al. (2018). Targeted delivery of a PD-1-blocking scFv by CAR-T cells enhances anti-tumor efficacy in vivo. Nat. Biotechnol. 36, 847–856. 10.1038/nbt.4195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Sharma P, Goswami S, Raychaudhuri D, Siddiqui BA, Singh P, Nagarajan A, Liu J, Subudhi SK, Poon C, Gant KL, et al. (2023). Immune checkpoint therapy-current perspectives and future directions. Cell 186, 1652–1669. 10.1016/j.cell.2023.03.006. [DOI] [PubMed] [Google Scholar]
- 283.Wykes MN, and Lewin SR (2018). Immune checkpoint blockade in infectious diseases. Nat. Rev. Immunol. 18, 91–104. 10.1038/nri.2017.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Bongartz T, Sutton AJ, Sweeting MJ, Buchan I, Matteson EL, and Montori V. (2006). Anti-TNF antibody therapy in rheumatoid arthritis and the risk of serious infections and malignancies: systematic review and meta-analysis of rare harmful effects in randomized controlled trials. JAMA 295, 2275–2285. 10.1001/jama.295.19.2275. [DOI] [PubMed] [Google Scholar]
- 285.Wenzel S, Ford L, Pearlman D, Spector S, Sher L, Skobieranda F, Wang L, Kirkesseli S, Rocklin R, Bock B, et al. (2013). Dupilumab in persistent asthma with elevated eosinophil levels. N. Engl. J. Med. 368, 2455–2466. 10.1056/NEJMoa1304048. [DOI] [PubMed] [Google Scholar]
- 286.Chan HT, Hughes D, French RR, Tutt AL, Walshe CA, Teeling JL, Glennie MJ, and Cragg MS (2003). CD20-induced lymphoma cell death is independent of both caspases and its redistribution into triton X-100 insoluble membrane rafts. Cancer Res. 63, 5480–5489. [PubMed] [Google Scholar]
- 287.Ichikawa K, Liu W, Zhao L, Wang Z, Liu D, Ohtsuka T, Zhang H, Mountz JD, Koopman WJ, Kimberly RP, and Zhou T. (2001). Tumoricidal activity of a novel anti-human DR5 monoclonal antibody without hepatocyte cytotoxicity. Nat. Med. 7, 954–960. 10.1038/91000. [DOI] [PubMed] [Google Scholar]
- 288.Przepiorka D, Ko CW, Deisseroth A, Yancey CL, Candau-Chacon R, Chiu HJ, Gehrke BJ, Gomez-Broughton C, Kane RC, Kirshner S, et al. (2015). FDA Approval: Blinatumomab. Clin. Cancer Res. 21, 4035–4039. 10.1158/1078-0432.CCR-15-0612. [DOI] [PubMed] [Google Scholar]
- 289.Dumontet C, Reichert JM, Senter PD, Lambert JM, and Beck A. (2023). Antibody-drug conjugates come of age in oncology. Nat. Rev. Drug Discov. 22, 641–661. 10.1038/s41573-023-00709-2. [DOI] [PubMed] [Google Scholar]
- 290.Saxton RA, Glassman CR, and Garcia KC (2023). Emerging principles of cytokine pharmacology and therapeutics. Nat. Rev. Drug Discov. 22, 21–37. 10.1038/s41573-022-00557-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Mo F, Yu Z, Li P, Oh J, Spolski R, Zhao L, Glassman CR, Yamamoto TN, Chen Y, Golebiowski FM, et al. (2021). An engineered IL-2 partial agonist promotes CD8+ T cell stemness. Nature 597, 544–548. 10.1038/s41586-021-03861-0. [DOI] [PMC free article] [PubMed] [Google Scholar]