Figure 2.
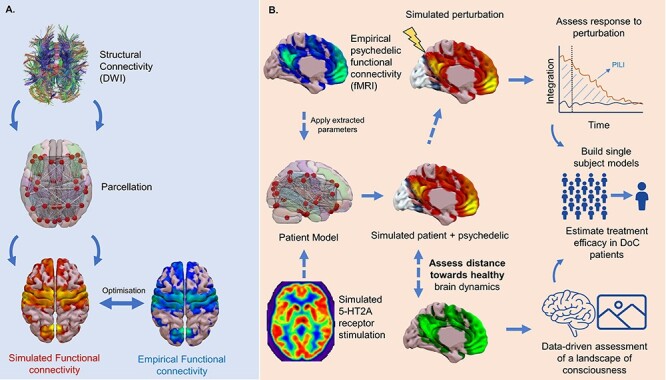
DoC = disorders of consciousness, PET = Positron emission tomography; 5-HT2A receptor= Serotonin 2A receptor; PILI= perturbative integration latency index; DWI = diffusion weighed imaging.
Illustration of the potential contribution of whole brain computational modelling. (a) The construction of a whole brain computational model from empirical structural connectivity derived from DWI. The model parameters are then optimised to achieve the best fit between simulated and empirical connectivity. (b) Parameters from models optimised on healthy controls under psychedelics could be extracted and applied to patient models to achieve a simulated psychedelic perturbation. Alternatively, neurotransmitter information from PET could be added to a dynamic mean field model to simulate receptor stimulation (i.e. serotonin-2A, 5-HT2A). The resulting dynamics could then be investigated through in silico perturbation protocols or through being compared to healthy subjects to assess the descriptive distance from the brains of healthy controls. Such techniques could build towards having single subject models through which psychedelic treatment could be initially assessed
