Figure 1.
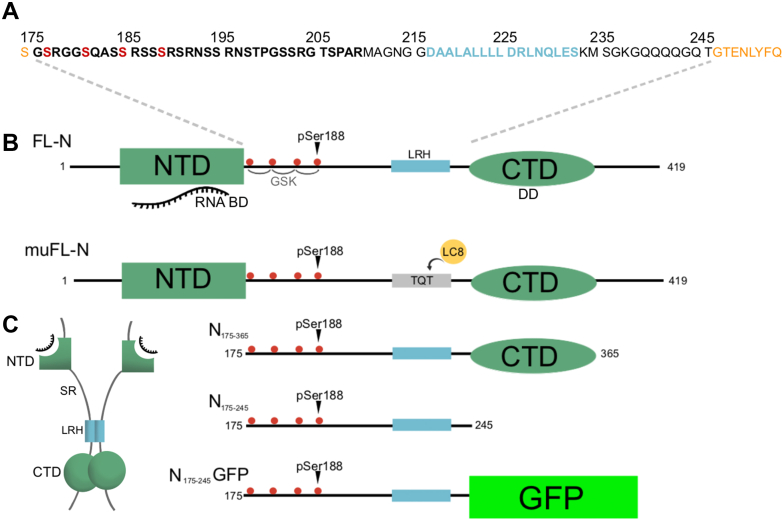
Domain maps of SARS-CoV-2N and constructs used in this work.A, amino acid sequence of SARS-CoV-2N construct spanning the linker region from residues 175 to 245. Residual TEV protease cleavage sites are in orange. The SR-rich region is in bold black, with phosphorylation sites investigated here in bold red. The Leu-rich helix (LRH) spanning residues 216 to 232 is in bold blue. B, domain maps of full-length (FL-N), mutant full-length (muFL-N), shorter constructs N175–365, N175–245, and GFP-tagged N175–245 from top to bottom, respectively. Intrinsically disordered regions are represented by black lines, the structured RNA-binding domain NTD is represented by a dark green rectangle, and the dimerization domain CTD by a dark green oval. The LRH is represented by a small blue rectangle. GSK priming site pSer188 is pointed to by an arrow, and the subsequent phosphorylation sites are indicated by red circles. The muFL-N has a TQT recognition motif for LC8-binding site incorporated to replace the LRH shown as gray rectangle. C, a cartoon depiction of dimeric FL-N showing self-association of LRH- and RNA-binding site of the NTD. CTD, C-terminal domain; GSK, glycogen synthase kinase; LC8, dynein light chain 8; LRH, Leu-rich helix; NTD, N-terminal domain; SR-rich, serine/arginine-rich.