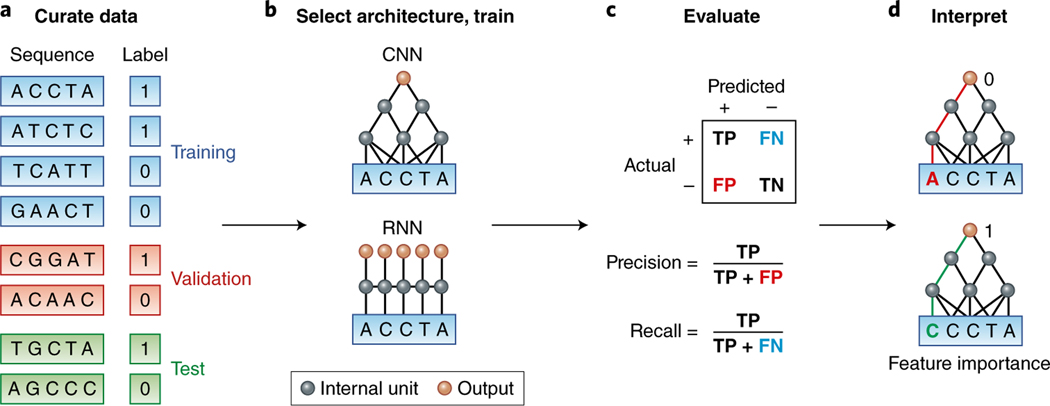
Fig. 1 |. Deep learning workflow in genomics.
a, A dataset should be randomly split into training, validation and test sets. The positive and negative examples should be balanced for potential confounders (for example, sequence content and location) so that the predictor learns salient features rather than confounders. b, The appropriate architecture is selected and trained on the basis of domain knowledge. For example, CNNs capture translation invariance, and RNNs capture more flexible spatial interactions. c, True positive (TP), false positive (FP), false negative (FN) and true negative (TN) rates are evaluated. When there are more negative than positive examples, precision and recall are often considered. d, The learned model is interpreted by computing how changing each nucleotide in the input affects the prediction. The interactive tutorial illustrates the four steps of this workflow (see URLs).