Abstract
Neuroimaging research requires purpose-built analysis software, which is challenging to install and may produce different results across computing environments. The community-oriented, open-source Neurodesk platform (https://www.neurodesk.org/) harnesses a comprehensive and growing suite of neuroimaging software containers. Neurodesk includes a browser-accessible virtual desktop, command-line interface and computational notebook compatibility, allowing for accessible, flexible, portable and fully reproducible neuroimaging analysis on personal workstations, high-performance computers and the cloud.
Neuroimaging data analysis is challenging. Aside from the scientific background motivating the choice of analysis, advanced domain knowledge beyond the researcher’s expertise is needed; for example, signal and image processing, software engineering, statistics and machine learning. Researchers faced with this task rely on specialized software packages typically developed by research teams with limited resources. The resulting analysis tools often have limited technical support, can be difficult to install, have conflicting dependencies or are inconsistently available across operating systems1–3. These issues not only are frustrating and time consuming, but also ultimately compromise reproducibility, a foundational scientific principle. We therefore developed Neurodesk, a community-oriented open-source solution for neuroimaging analysis with four guiding principles: accessibility, portability, flexibility and, overarchingly, reproducibility.
Ideally, scientific analysis workflows should be easily accessible, so users can deploy them from any computing environment with minimal time and effort4, and portable, so that users can tractably shift analysis pipelines between computing environments once developed. Many researchers prototype analysis pipelines using their laptop or desktop computer, and then switch to workstations and high-performance computing clusters for processing at scale. Accessible and portable workflows allow for the optimized allocation of computing resources while supporting shared development workloads5. Unfortunately, many analysis workflows are neither readily accessible nor portable6,7, and many existing solutions to these issues lack flexibility8. For example, single-install preprogrammed analysis pipelines are popular with clinicians, but researchers typically customize analysis pipelines for specific projects9–11. Virtual machines or dual-boot computers partially address these barriers, but they are resource intensive and still do not reconcile conflicts between software packages or their dependencies. Beyond productivity costs, inaccessible and unstable neuroimaging tools also pose a wider threat to reproducibility12,13, that is, running the same software on the same input data and obtaining the same result14. The transparency and openness promotion guidelines, which have over 5,000 journals, publishers and other related organizations as signatories, state that all reported results should be independently reproduced before publication15. But realistically, results verification is usually too impractical to implement at review6.
These issues are not unique to neuroimaging or scientific research, and similar issues in the software space led to the development of software containers; lightweight and portable solutions that package applications and their dependencies. Container engines such as Docker, Podman and Apptainer/Singularity allow containerized software to seamlessly shift between computing environments without relying on, or conflicting with, software outside the container16. Containers are thus well suited to address the issues facing neuroimaging analysis and form the core of the Neurodesk project17 (Fig. 1). Neurodesk makes containerized neuroimaging software easier to both access and create through the Neurocontainers repository, a comprehensive and growing collection of versioned neuroimaging software containers (Fig. 1a,b). Neurocontainers contributed by the community are automatically made available to access through Neurodesk (Fig. 1a). Each Neurocontainer includes the packaged tool and all dependencies required to execute a specific version of that tool (Fig. 1c). Because containers isolate dependencies, different Neurocontainers can provide different versions of the same tool, allowing researchers to seamlessly switch software versions.
Fig. 1 |. The Neurodesk platform.
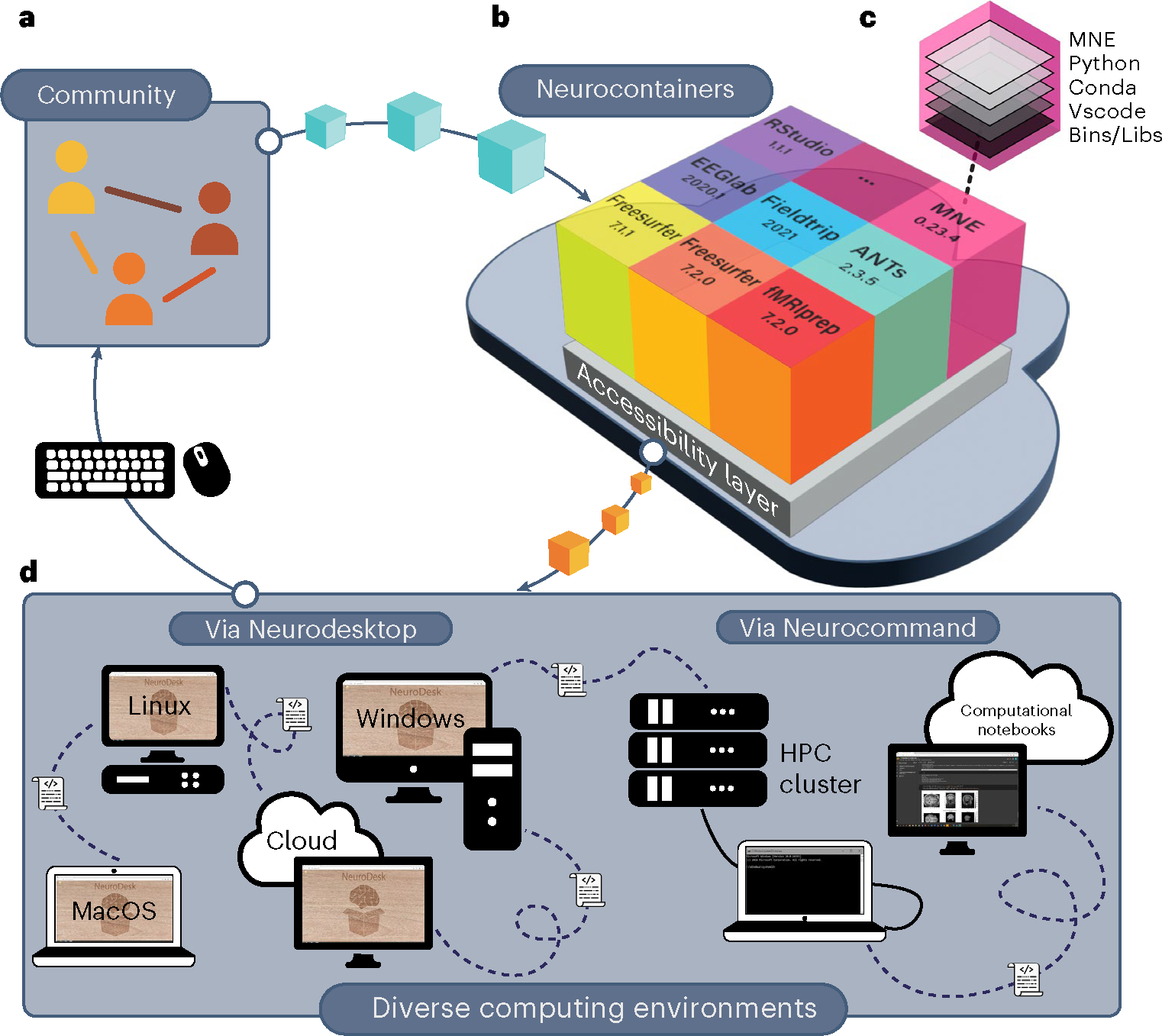
a, Neurodesk is built by and for the scientific community, enabling anyone to contribute containers. b, Community-contributed software recipes are automatically used to build software containers stored in the Neurocontainers repository. c, Each container packages a tool together with all its dependencies. d, Neurodesk provides two layers of accessibility: (1) Neurodesktop: a browser-accessible virtual desktop environment; (2) Neurocommand: a command-line interface that runs the same software containers programmatically. These interfaces allow users to reproduce analyses across computing environments (HPC: high-performance computing).
Neurodesk enables researchers to use Neurocontainers directly through the cloud or download containers for offline use through two possible interfaces, without the need to install neuroimaging software locally. First, Neurodesktop is a browser-accessible virtual desktop environment with all containerized tools accessible from the application menu (Fig. 1d). Neurodesktop has the look and feel of working on one’s local computer, and can be executed using local or cloud resources. Second, Neurocommand can be used to launch and interact with Neurocontainers through the command line. Neurocommand is suitable for use in high-performance computing environments, and can be used to interact with neuroimaging software through computational notebooks such as Google Colab or Jupyter Notebooks18 (Fig. 1d). These Neurodesk interfaces can be launched from most common operating systems by installing the Neurodesk App, or by launching remote instances online. Extensive documentation, tutorials and examples are available at the Neurodesk website (https://www.neurodesk.org/). By harnessing these easy-to-use interfaces, researchers can flexibly take advantage of large open datasets, reproduce reported analyses, and switch between neuroimaging modalities and computational platforms within and between projects. Containerized software reduces unnecessary computational variability between execution systems, making it possible to share analyses between laboratories and collaborate on large datasets without artificial differences between sites. Further, for developers, the effort to containerize and add one’s software to Neurodesk may be minimal compared with testing software and supporting users across diverse computing platforms.
Studies have shown that subtle differences in hardware, firmware and software dependencies can systematically alter results across computing environments19–21, meaning it is often impossible to replicate results even when given the original data, code and software version. This effect has been well described for functional magnetic resonance imaging (fMRI) processing pipelines19. To evaluate whether Neurodesk addresses these issues, we therefore set out to replicate and extend upon these findings; we ran four identical MRI analysis pipelines, in two separate computing environments, using software installed locally and through Neurodesk. We found meaningful differences in image intensity and subcortical tissue classification between the two computers for pipelines run on locally installed software (Fig. 2a,b), but not for pipelines run on Neurodesk (Fig. 2c,d). These results show that Neurodesk allows researchers to adhere to the highest possible reproducibility standards with minimal changes to their typical workflow. See the Supplementary Notes for the full results of this case study.
Fig. 2 |. Inter-computer differences in an fMRI processing pipeline.
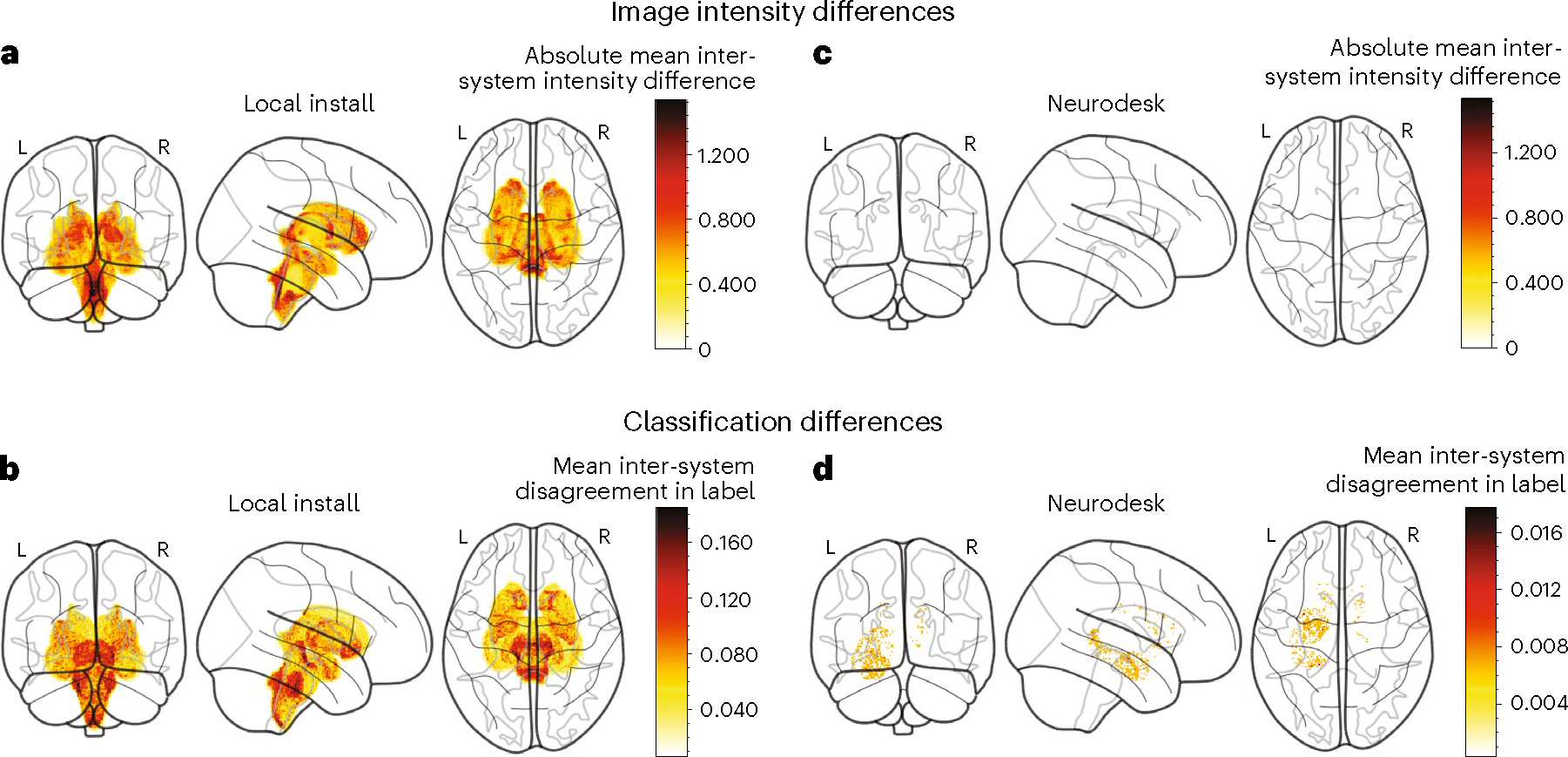
a,c, Absolute mean inter-computer image intensity differences within subcortical structures after image registration with FSL-FLIRT. Projections are shown for locally installed software (a) and Neurodesk (c). b,d, Inter-system classification disagreement after image segmentation with FSL-FIRST, averaged across participants. Projections are shown for locally installed software (b) and Neurodesk (d) (note the difference in color scale range).
Neurodesk not only facilitates access to reproducible neuroimaging data analysis, but also makes sharing these workflows less burdensome. Neurocontainers are accessible within computational notebooks (for example, running FreeSurfer22 within Google Colab), enabling researchers to share reproducible code and results alongside published manuscripts. Notably, this approach requires authors to ensure interoperability of the linked code and data, ensuring that readers do not need to spend time downloading large datasets from remote repositories, or overcome issues with executing notebooks due to insufficient cloud computing resources. Recent developments in reproducible preprints present an enriched publication path that simplifies the sharing of data and analysis code23. NeuroLibre, for example, hosts interactive notebooks and associated data, allowing readers to modify and re-execute code24. Neurocontainers are ideally suited for such integrated and reproducible approaches.
Neurodesk is also impactful as an educational tool in workshops and courses. The platform was first conceptualized during a ‘hackathon’25, an event where people with diverse skill sets collaborated on projects and developed research skills. Variability in analysis environments across attendees’ computers presents a hurdle for neuroimaging training workshops such as this. Facilitators often spend considerable time troubleshooting software installations specific to unique computing environments. Neurodesk, which provides access to a standardized analysis environment with the requisite tools preinstalled with almost no set up, allows researchers to efficiently tackle complex scientific problems by eliminating technical troubleshooting. Moreover, Neurodesk is scalable to different class sizes and computational demands, can be accessed remotely and enables trainees to easily access their analyses after the workshop. Containerized platforms in other fields have made a substantial impact in this way, for example, the Galaxy platform for bioinformatics26.
Neurodesk exists within a larger ecosystem of projects providing accessible, reproducible, flexible and portable neuroimaging analysis, and, where possible, seeks to interoperate with related platforms. While Neurodesk is not the only project to address any one of these principles, Neurodesk is unique in addressing all four principles. Projects such as NeuroDebian1 and Neurofedora27 increase accessibility for GNU/Linux operating systems, but offer limited support for portability or reproducibility. Other projects such as Brainlife28, BIDSApps29, Flywheel (https://flywheel.io/), XNAT30, Code-Ocean31, Qmenta (https://www.qmenta.com/), CBRAIN32 and Biocontainers33 all support reproducibility through containerization, but have different use-cases to Neurodesk. For example, Brainlife facilitates reproducible and traceable cloud-based analysis using community-contributed workflows. However, the platform is designed to allow users to run pre-coded analysis pipelines, rather than to flexibly access software to develop their own pipelines. To this end, the Neurodesk and Brainlife teams are increasing interoperability between the platforms by providing Brainlife development environments on Neurodesk, and running Neurodesk containers on Brainlife. Thus, in cases where flexibility is less important, Neurodesk can also be harnessed to support the complete workflow reproducibility offered by preprogrammed analysis pipelines with Brainlife. Similarly, we have integrated the ‘BIDSApps’ repository of containerized workflows, allowing users to access or adapt these workflows. In this way, users already accustomed to other platforms and tools will also benefit from the Neurodesk project.
Neurodesk has some limitations that warrant discussion. One area of ongoing development relates to the inclusion of proprietary and licensed software without compromising accessibility. Another challenge for a flexible platform with as wide a range of applications as Neurodesk is the project’s long-term sustainability. Neurodesk’s community-driven, continuous integration model provides a powerful and flexible way to address both of these expanded use-cases without depending on a single development team. We have developed multiple pathways for sustainability, including the federated support of the underlying hosting infrastructure, flexibility in the continuous integration and deployment infrastructure and a potential for a commercial model to offer tailored support for institutions and workshops.
The challenges of accessibility, portability, flexibility and reproducibility discussed here are not unique to neuroscience. In turn, Neurodesk’s core foundation could be used to deploy software specific to any other discipline, and it is our sincere hope that this platform is adapted as such. The Neurodesk platform has the potential to improve the way scientists analyze data and communicate results. Specifically, Neurodesk allows any scientist, anywhere in the world, to conveniently access, develop and adapt their neuroimaging analysis tools, and apply them in a fully reproducible manner from any computing environment.
Online content
Any methods, additional references, Nature Portfolio reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at https://doi.org/10.1038/s41592-023-02145-x.
Methods
How to use Neurodesk: accessibility, flexibility and portability
The Neurodesk platform’s website (https://www.neurodesk.org/) is user-friendly and open to community contributions. The website contains information about the included software and is automatically updated through continuous integration. Therefore, there is always up-to-date documentation, lists of currently available applications and a release history. The website also hosts clear instructions for accessing and interacting with Neurodesk from various computing environments and tutorials on using various software packages.
Neurodesk makes reproducible neuroimaging data analysis accessible in almost any computing environment and brings the same dependencies to all supported platforms. This portability extends to the Neurodesktop graphical user interface, which provides the same desktop environment across all supported computing environments. Containerized analyses look, feel and run the same way across different computing environments. Thus, researchers reading or reviewing manuscripts with open data and code can use Neurodesk to replicate the exact pipeline using the reported tool versions without being required to install additional software.
For a data analysis environment to be portable, such that it can easily shift between computing environments, it also needs to be lightweight with a small storage footprint. To this end, our accessibility layer harnesses the CernVM File System (CVMFS)34. The CVMFS layer allows accessing the software from a remote host without installation, so only parts of a container that are actively used are sent over the network and cached on the user’s local computer. Users can access terabytes of software without explicitly downloading or storing it locally. The Neurodesk platform has several CVMFS nodes worldwide, providing low latency and direct access to Neurocontainers. Thus, to use Neurodesk, users only install the required container engine to access the Neurocontainer of their choice. The current release of Neurodesktop, which facilitates access to all tools in the Neurocontainers repository, is less than 1.6 GB in download size.
Anticipating that installing a third-party container engine software may be a barrier to entry for some researchers, there is an entirely cloud-based solution: ‘Neurodesk Play’ (https://play.neurodesk.org/). Neurodesk Play is accessible globally, allowing anyone to use a cloud-based graphical desktop environment for neuroimaging data analysis and teaching. Neurodesk play instances are Binderhub35 instances deployed based on the zero-to-binderhub guide, coupled with the full suite of Neurocontainers delivered via CVMFS. Neurodesktop can also run on institutional or cloud computing resources enabling access to large amounts of computing resources or datasets. For example, the Australian Research Data Commons (ARDC) provides Neurodesk on their Virtual Desktop Service freely available to anyone with an Australian Access Federation account.
The accessibility, flexibility and portability of this platform can be best assessed through its utility to users. We, therefore, display up-to-date usage statistics for the platform on our website (https://www.neurodesk.org/docs/overview/metrics/). Further, the platform has already been referenced in several peer-reviewed studies36–39.
Long-term sustainability of the Neurodesk platform
Neurodesk has a wide selection of tools available spanning many domains of neuroimaging data analysis. Extended Data Table 1 shows the tools available at the time of publication, although this list is growing rapidly as the community and developers contribute software through recipes created using the open-source Neurodocker project40. These recipes can be based on the Neurodebian project1. Users can find a full and up-to-date list at https://www.neurodesk.org/applications/. Neurodesk uses a two-pronged approach to staying up to date with new neuroimaging tools and new versions of already included software: (1) The Neurodesk maintainers add tools as they become aware of new developments or community members request the addition of new packages. The Neurodesk GitHub organization (https://github.com/NeuroDesk/) has an active discussion forum where developers respond to requests for new software containers. (2) In addition to this developer-centric route to new software containers, we actively encourage contributions from the research community. A core aim for developing the Neurodesk platform was to build it as a community-driven project that is not contingent on a specific team of developers. As such, we provide a template and detailed instructions for creating build scripts for new software containers. Moreover, we aim to ensure long-term executability of the containers by storing the containers in different formats: docker, podman, singularity/apptainer and an unpacked chroot environment. This comes with the benefit of increased accessibility for users and the advantage that when technology progresses and standards change over the years, users will still be able to execute the software through standard GNU/Linux kernel tools (chroot and mount)41–99.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Extended Data
Extended Data Table 1.
Tools currently available in Neurodesk
| Category | Tool |
|---|---|
| Editors and Programming | VS Code, Gedit, Emacs, Vim, Python, Git, Julia, Matlab, ROOT, RStudio |
| Data Synchronisation Tools | Rsync, Rclone, Nextcloud client, Owncloud client, Globus personal connect |
| Workflows | Nipype42, ASLPrep43, fMRIPrep44, MRIQC45, QSMxT39 |
| Data Organisation | dcm2niix46, BIDScoin47, BIDStools48, Convert3D49 |
| Diffusion MRI | Diffusion Toolkit50, DSI Studio51, MRtrix52, MRtrix3Tissue (www.3Tissue.github.io), TrackVis50 |
| Rodent Imaging | AIDAmri53, RABIES54 |
| Spectroscopy | LCModel (http://s-provencher.com/lcmodel.shtml), MRSIProc55 |
| Structural and/or Functional Imaging | AFNI56, ANTS57, ASHS58, BART (https://mrirecon.github.io/bart/), CAT1259, CLEAR-SWI60, Conn61, Connectome Workbench62, FatSegNet63, FreeSurfer22, FSL64, HD-BET65, LASHiS66, LayNii67, MINC68, MRItools69, NiftyReg (https://www.nitrc.org/projects/niftyreg/), NiiStat (https://www.nitrc.org/projects/niistat/), OSHy-X38, Palm Alpha70, PhysIO71, ROMEO72, Slicer73, Spinal Cord Toolbox74, SPM75, TGVQSM76, elastix77,78, mfcsc79 |
| Electroencephalography (EEG)and/or Magnetoencephalography (MEG) | Brainstorm80, EEGLAB81, FieldTrip82, MNE83, Sigviewer84 |
| Machine Learning and Statistics | R85, Deep Retinopy86, Delphi87 |
| Visualisation and Image Editing | ImageMagick88, GIMP (www.gimp.org), itk-SNAP49, MRIcron89, MRIcroGL90, SicerSALT91, Surf Ice92, VesselVio93 |
| BIDS App | Automatic Analysis94, BARACUS95, BrainSuite96, HCPPipelines97,98, MRtrix3_connectome (https://github.com/bids-apps/MRtrix3_connectome) |
| Molecular biology | MGLTools (https://ccsb.scripps.edu/mgltools/), AutoDock Vina99,100 |
The Neurodesk development team uses a broad definition of what constitutes a ‘tool’ and is guided by the community in what level of granularity would most flexibly facilitate neuroimaging data analysis on a case-by-case basis. Note that each tool has been listed under only one category, although some may span multiple categories. An up-to-date table can be retrieved from https://www.neurodesk.org/applications/. Details on the tools are available in refs. 41–99.
Supplementary Material
Acknowledgements
The ARDC invested in Neurodesk’s development through the Australian Electrophysiology Data Analytics Platform project (S.B., A.N., O.C., T.J. and R.S.). We thank Oracle for Research for providing Oracle Cloud credits and related cloud resources to support this project (S.B.) The University of Queensland funded the project via the Knowledge Exchange & Translation Fund and the UQ AI Collaboratory (S.B.). S.B., F.L.R. and A.W.S. acknowledge funding through an ARC Linkage grant (LP200301393). S.B. and A.W.S. acknowledge funding through the Australian Research Council Training Centre for Innovation in Biomedical Imaging Technology (IC170100035). This research was supported by use of the Nectar Research Cloud, a collaborative Australian research platform supported by the National Collaborative Research Infrastructure Strategy-funded ARDC. We acknowledge the facilities and scientific and technical assistance of the National Imaging Facility, a National Collaborative Research Infrastructure Strategy capability. A National Institutes of Health grant (P41EB019936) partially supported J.R.K. and S.S.G. Data collection and sharing for this project was provided by the International Consortium for Brain Mapping (ICBM; Principal Investigator: J. Mazziotta). ICBM funding was provided by the National Institute of Biomedical Imaging and BioEngineering. ICBM data are disseminated by the Laboratory of Neuro Imaging at the University of Southern California. We thank I. C. D. Lenton, E. Cooper-Williams and Y. ‘Sam’ Peng for contributions to the first NeuroDesk precursor ‘Dicom2Cloud’ and the reviewers for the constructive feedback. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Footnotes
Competing interests
The authors declare no competing interests.
Code availability
The code for this project is publicly available on GitHub, across multiple repositories under the https://github.com/NeuroDesk/ organization. It has also been archived on Zenodo at https://doi.org/10.5281/zenodo.8053090. The code is licensed under the MIT License.
All stages of development, from the initial conception as a hackathon project, through to the most current iteration of Neurodesk, with up-to-date community-built Neurocontainer recipes, are documented publicly across the project’s GitHub repository and the platform’s website; which contains descriptions of how code is organized on the GitHub repository, and how to contribute to the project (https://www.neurodesk.org/).
Any issues can be logged at https://github.com/orgs/NeuroDesk/discussions/. Contributions can be made by any community member with a GitHub account and the eagerness to create pull requests.
Additional information
Extended data is available for this paper at https://doi.org/10.1038/s41592-023-02145-x.
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41592-023-02145-x.
Peer review information Nature Methods thanks Taiga Abe, Agah Karakuzu, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Nina Vogt, in collaboration with the Nature Methods team. Peer reviewer reports are available.
Reprints and permissions information is available at www.nature.com/reprints.
Data availability
The data that support the findings of the case study are available from the ICBM database (https://www.loni.usc.edu/). There are restrictions that apply to the availability of these data, which were used under approved permission for the current study, and thus are not publicly available but are available from ICBM upon request. Source data are provided with this paper.
References
- 1.Halchenko Y & Hanke M Open is not enough. let’s take the next step: an integrated, community-driven computing platform for neuroscience. Front. Neuroinform. 6, 22 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hanke M & Halchenko Y Neuroscience runs on GNU/Linux. Front. Neuroinform. 5, 8 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Niso G et al. Open and reproducible neuroimaging: from study inception to publication. NeuroImage 263, 119623 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilkinson MD et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data 3, 160018 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kurtzer GM, Sochat V & Bauer MW Singularity: scientific containers for mobility of compute. PLoS ONE 12, e0177459 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van Gorp P & Mazanek S SHARE: a web portal for creating and sharing executable research papers. Procedia Comput. Sci. 4, 589–597 (2011). [Google Scholar]
- 7.Poline J-B. et al. Is neuroscience FAIR? a call for collaborative standardisation of neuroscience data. Neuroinformatics 20, 507–512 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Silberzahn R et al. Many analysts, one data set: making transparent how variations in analytic choices affect results. Adv. Methods Pract. Psychol. Sci. 1, 337–356 (2018). [Google Scholar]
- 9.Tapera TM et al. FlywheelTools: data curation and manipulation on the Flywheel platform. Front. Neuroinform. 15, 678403 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Routier A et al. Clinica: an open-source software platform for reproducible clinical neuroscience studies. Front. Neuroinform. 15, 689675 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abe T et al. Neuroscience cloud analysis as a service: an open-source platform for scalable, reproducible data analysis. Neuron 110, 2771–2789 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goodman SN, Fanelli D & Ioannidis JPA What does research reproducibility mean? Sci. Transl. Med. 8, 341ps12–341ps12 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Nosek BA et al. Replicability, robustness, and reproducibility in psychological science. Annu. Rev. Psychol. 73, 719–748 (2022). [DOI] [PubMed] [Google Scholar]
- 14.Plesser HE Reproducibility vs. replicability: a brief history of a confused terminology. Front. Neuroinform. 11, 76 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nosek BA et al. Promoting an open research culture. Science 348, 1422–1425 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boettiger C An introduction to Docker for reproducible research. ACM SIGOPS Oper. Syst. Rev. 49, 71–79 (2015). [Google Scholar]
- 17.Trunov AS, Voronova LI, Voronov VI & Ayrapetov DP Container cluster model development for legacy applications integration in scientific software system. in 2018 IEEE International Conference ‘Quality Management, Transport and Information Security, Information Technologies’ (IT QM IS) 815–819 10.1109/ITMQIS.2018.8525120 (2018). [DOI] [Google Scholar]
- 18.Thomas T et al. Jupyter Notebooks—a publishing format for reproducible computational workflows. In Positioning and Power in Academic Publishing: Players, Agents and Agendas (eds Loizides F. & Schmid B.) 87–90 (IOS Press, 2016). [Google Scholar]
- 19.Glatard T et al. Reproducibility of neuroimaging analyses across operating systems. Front. Neuroinform. 9, 12 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gronenschild EH et al. The effects of FreeSurfer version, workstation type, and Macintosh operating system version on anatomical volume and cortical thickness measurements. PLoS ONE 7, e38234 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krefting D et al. Reliability of quantitative neuroimage analysis using freesurfer in distributed environments. In MICCAI Workshop on High-Performance and Distributed Computing for Medical Imaging (2011). [Google Scholar]
- 22.Fischl B FreeSurfer. NeuroImage 62, 774–781 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.DuPre E et al. Beyond advertising: new infrastructures for publishing integrated research objects. PLoS Comput. Biol. 18, e1009651 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karakuzu A et al. NeuroLibre: a preprint server for full-fledged reproducible neuroscience. Preprint at OSF 10.31219/osf.io/h89js (2022). [DOI] [Google Scholar]
- 25.Gau R et al. Brainhack: developing a culture of open, inclusive, community-driven neuroscience. Neuron 109, 1769–1775 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Afgan E et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 46, W537–W544 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sinha A et al. Comp-NeuroFedora, a free/open source operating system for computational neuroscience: download, install, research. BMC Neurosci. 21, 1 (2020).31941442 [Google Scholar]
- 28.Hayashi S et al. brainlife.io: a decentralized and open source cloud platform to support neuroscience research. Preprint at arXiv 10.48550/arXiv.2306.02183 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gorgolewski KJ et al. BIDS apps: improving ease of use, accessibility, and reproducibility of neuroimaging data analysis methods. PLoS Comput. Biol. 13, e1005209 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Herrick R et al. XNAT Central: open sourcing imaging research data. NeuroImage 124, 1093–1096 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Staubitz T, Klement H, Teusner R, Renz J & Meinel C CodeOcean—a versatile platform for practical programming excercises in online environments. In 2016 IEEE Global Engineering Education Conference (EDUCON) 314–323 10.1109/EDUCON.2016.7474573 (2016). [DOI] [Google Scholar]
- 32.Sherif T et al. CBRAIN: a web-based, distributed computing platform for collaborative neuroimaging research. Front. Neuroinform. 8, 54 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.da Veiga Leprevost F et al. BioContainers: an open-source and community-driven framework for software standardization. Bioinformatics 33, 2580–2582 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blomer J et al. Micro-CernVM: slashing the cost of building and deploying virtual machines. J. Phys. Conf. Ser. 513, 032009 (2014). [Google Scholar]
- 35.Jupyter P et al. Binder 2.0—reproducible, interactive, sharable environments for science at scale. in Proceedings of the 17th Python in Science Conference 113–120 10.25080/Majora-4af1f417-011 (2018). [DOI] [Google Scholar]
- 36.Atilgan H et al. Functional relevance of the extrastriate body area for visual and haptic object recognition: a preregistered fMRI-guided TMS study. Cereb. Cortex Commun. 4, tgad005 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang J et al. Open-source hypothalamic-ForniX (OSHy-X) atlases and segmentation tool for 3T and 7T. J. Open Source Softw. 7, 4368 (2022). [Google Scholar]
- 38.Stewart AW et al. QSMxT: robust masking and artifact reduction for quantitative susceptibility mapping. Magn. Reson. Med. 87, 1289–1300 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Biondetti E et al. Multi-echo quantitative susceptibility mapping: how to combine echoes for accuracy and precision at 3 Tesla. Magn. Reson. Med. 88, 2101–2116 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaczmarzyk J et al. ReproNim/neurodocker: 0.9.5. 10.5281/zenodo.7929032 (2023). [DOI] [Google Scholar]
- 41.Gorgolewski K et al. Nipype: a flexible, lightweight and extensible neuroimaging data processing framework in Python. Front. Neuroinform. 5, 13 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adebimpe A et al. ASLPrep: a platform for processing of arterial spin labeled MRI and quantification of regional brain perfusion. Nat. Methods 19, 683–686 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Esteban O et al. fMRIPrep: a robust preprocessing pipeline for functional MRI. Nat. Methods 16, 111–116 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Esteban O et al. MRIQC: Advancing the automatic prediction of image quality in MRI from unseen sites. PLoS ONE 12, e0184661 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li X, Morgan PS, Ashburner J, Smith J & Rorden C The first step for neuroimaging data analysis: DICOM to NIfTI conversion. J. Neurosci. Methods 264, 47–56 (2016). [DOI] [PubMed] [Google Scholar]
- 46.Zwiers MP, Moia S & Oostenveld R BIDScoin: a user-friendly application to convert source data to brain imaging data structure. Front. Neuroinform. 15, 770608 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gorgolewski KJ et al. The brain imaging data structure, a format for organizing and describing outputs of neuroimaging experiments. Sci. Data 3, 160044 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yushkevich PA et al. User-guided segmentation of multi-modality medical imaging datasets with ITK-SNAP. Neuroinformatics 17, 83–102 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang R, Benner T, Sorensen AG & Wedeen VJ Diffusion toolkit: a software package for diffusion imaging data processing and tractography. Proc. Intl Soc. Mag. Reson. Med. 15, 3720 (2007). [Google Scholar]
- 50.Yeh F-C. Population-based tract-to-region connectome of the human brain and its hierarchical topology. Nat. Commun. 13, 4933 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tournier J-D., Calamante F. & Connelly A. MRtrix: diffusion tractography in crossing fiber regions. Int. J. Imaging Syst. Technol. 22, 53–66 (2012). [Google Scholar]
- 52.Pallast N et al. Processing pipeline for atlas-based imaging data analysis of structural and functional mouse brain MRI (AIDAmri). Front. Neuroinform. 13, 42 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Desrosiers-Gregoire G et al. Rodent Automated Bold Improvement of EPI Sequences (RABIES): a standardized image processing and data quality platform for rodent fMRI. Preprint at bioRxiv 10.1101/2022.08.20.504597 (2022). [DOI] [Google Scholar]
- 54.Hangel G et al. Ultra-high resolution brain metabolite mapping at 7T by short-TR Hadamard-encoded FID-MRSI. NeuroImage 168, 199–210 (2018). [DOI] [PubMed] [Google Scholar]
- 55.Cox RW AFNI: what a long strange trip it’s been. NeuroImage 62, 743–747 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Avants BB, Tustison N & Johnson H Advanced Normalization Tools (ANTS). Insight J. 2, 1–35 (2009). [Google Scholar]
- 57.Wisse LEM et al. Automated hippocampal subfield segmentation at 7T MRI. Am. J. Neuroradiol. 37, 1050–1057 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gaser C et al. CAT—a computational anatomy toolbox for the analysis of structural MRI data. Preprint at bioRxiv 10.1101/2022.06.11.495736 (2022). [DOI] [Google Scholar]
- 59.Eckstein K et al. Improved susceptibility weighted imaging at ultra-high field using bipolar multi-echo acquisition and optimized image processing: CLEAR-SWI. NeuroImage 237, 118175 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Whitfield-Gabrieli S & Nieto-Castanon A Conn: a functional connectivity toolbox for correlated and anticorrelated brain networks. Brain Connect 2, 125–141 (2012). [DOI] [PubMed] [Google Scholar]
- 61.Marcus DS et al. Human Connectome Project informatics: quality control, database services, and data visualization. NeuroImage 80, 202–219 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Estrada S et al. FatSegNet: a fully automated deep learning pipeline for adipose tissue segmentation on abdominal dixon MRI. Magn. Reson. Med. 83, 1471–1483 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jenkinson M, Beckmann CF, Behrens TEJ, Woolrich MW & Smith SM FSL. NeuroImage 62, 782–790 (2012). [DOI] [PubMed] [Google Scholar]
- 64.Isensee F et al. Automated brain extraction of multisequence MRI using artificial neural networks. Hum. Brain Mapp. 40, 4952–4964 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shaw T, York A, Ziaei M, Barth M & Bollmann S Longitudinal Automatic Segmentation of Hippocampal Subfields (LASHiS) using multi-contrast MRI. NeuroImage 218, 116798 (2020). [DOI] [PubMed] [Google Scholar]
- 66.Huber LR et al. LayNii: a software suite for layer-fMRI. NeuroImage 237, 118091 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vincent RD et al. MINC 2.0: a flexible format for multi-modal images. Front. Neuroinformatics 10, 35 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Grussu F et al. Multi-parametric quantitative in vivo spinal cord MRI with unified signal readout and image denoising. NeuroImage 217, 116884 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Winkler AM, Ridgway GR, Webster MA, Smith SM & Nichols TE Permutation inference for the general linear model. NeuroImage 92, 381–397 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kasper L et al. The PhysIO toolbox for modeling physiological noise in fMRI data. J. Neurosci. Methods 276, 56–72 (2017). [DOI] [PubMed] [Google Scholar]
- 71.Dymerska B et al. Phase unwrapping with a rapid opensource minimum spanning tree algorithm (ROMEO). Magn. Reson. Med. 85, 2294–2308 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fedorov A et al. 3D slicer as an image computing platform for the quantitative imaging network. Magn. Reson. Imaging 30, 1323–1341 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.De Leener B et al. SCT: Spinal Cord Toolbox, an open-source software for processing spinal cord MRI data. NeuroImage 145, 24–43 (2017). [DOI] [PubMed] [Google Scholar]
- 74.Ashburner J Computational anatomy with the SPM software. Magn. Reson. Imaging 27, 1163–1174 (2009). [DOI] [PubMed] [Google Scholar]
- 75.Langkammer C et al. Fast quantitative susceptibility mapping using 3D EPI and total generalized variation. NeuroImage 111, 622–630 (2015). [DOI] [PubMed] [Google Scholar]
- 76.Klein S, Staring M, Murphy K, Viergever MA & Pluim J elastix: a toolbox for intensity-based medical image Registration. IEEE Trans. Med. Imaging 29, 196–205 (2010). [DOI] [PubMed] [Google Scholar]
- 77.Shamonin D et al. Fast parallel image registration on CPU and GPU for diagnostic classification of Alzheimer’s disease. Front. Neuroinform. 7, 50 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Civier O, Sourty M & Calamante F MFCSC: novel method to calculate mismatch between functional and structural brain connectomes, and its application for detecting hemispheric functional specialisations. Sci. Rep. 13, 3485 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tadel F, Baillet S, Mosher JC, Pantazis D & Leahy RM Brainstorm: a user-friendly application for MEG/EEG analysis. Comput. Intell. Neurosci. 2011, 879716 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Brunner C, Delorme A & Makeig S Eeglab—an open source MATLAB toolbox for electrophysiological research. Biomed. Tech. 58, 1 (2013). [DOI] [PubMed] [Google Scholar]
- 81.Oostenveld R, Fries P, Maris E & Schoffelen J-M. FieldTrip: open source software for advanced analysis of MEG, EEG, and invasive electrophysiological data. Comput. Intell. Neurosci. 2011, 156869 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gramfort A et al. MNE software for processing MEG and EEG data. NeuroImage 86, 446–460 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Brunner C, Breitwieser C & Müller-Putz GR Sigviewer and Signalserver—open source software projects for biosignal analysis. Biomed. Eng. Tech. 58, 1 (2013). [DOI] [PubMed] [Google Scholar]
- 84.Ihaka R & Gentleman RR: a language for data analysis and graphics. J. Comput. Graph. Stat. 5, 299–314 (1996). [Google Scholar]
- 85.Ribeiro FL, Bollmann S & Puckett AM Predicting the retinotopic organization of human visual cortex from anatomy using geometric deep learning. NeuroImage 244, 118624 (2021). [DOI] [PubMed] [Google Scholar]
- 86.Mishra P, Lehmkuhl R, Srinivasan A, Zheng W & Popa RA Delphi: a cryptographic inference service for neural networks. In 29th USENIX Security Symposium (USENIX Security 20) 2505–2522 (2020). [Google Scholar]
- 87.Still M The definitive guide to ImageMagick. vol. 1 (Springer, 2006). [Google Scholar]
- 88.Rorden C & Brett M Stereotaxic display of brain lesions. Behav. Neurol. 12, 191–200 (2000). [DOI] [PubMed] [Google Scholar]
- 89.Rorden C rordenlab/MRIcroGL: version 20-July-2022 (v1.2.20220720) 10.5281/ZENODO.7533834 (2022). [DOI] [Google Scholar]
- 90.Vicory J et al. SlicerSALT: Shape AnaLysis Toolbox. In Shape in Medical Imaging (eds. Reuter M. et al.) vol. 11167, 65–72 (Springer International Publishing, 2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rorden C & Hanayik T neurolabusc/surf-ice: version 6-October-2021 (v1.0.20211006). 10.5281/ZENODO.7533772 (2021) [DOI] [Google Scholar]
- 92.Bumgarner JR & Nelson RJ Open-source analysis and visualization of segmented vasculature datasets with VesselVio. Cell Rep. Methods 2, 100189 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cusack R et al. Automatic analysis (aa): efficient neuroimaging workflows and parallel processing using Matlab and XML. Front. Neuroinform. 8, 90 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Liem F & Gorgolewski CF BIDS-Apps/baracus: v1.1.2. 10.5281/ZENODO.1018841 (2017). [DOI] [Google Scholar]
- 95.Kim Y et al. BrainSuite BIDS App: containerized workflows for MRI analysis. Preprint at bioRxiv 10.1101/2023.03.14.532686 (2023). [DOI] [Google Scholar]
- 96.Glasser MF et al. The minimal preprocessing pipelines for the Human Connectome Project. NeuroImage 80, 105–124 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Smith SM et al. Resting-state fMRI in the Human Connectome Project. NeuroImage 80, 144–168 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Trott O & Olson AJ AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, 455–461 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Eberhardt J, Santos-Martins D, Tillack AF & Forli S AutoDock Vina 1.2.0: new docking methods, expanded force field, and Python bindings. J. Chem. Inf. Model. 61, 3891–3898 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of the case study are available from the ICBM database (https://www.loni.usc.edu/). There are restrictions that apply to the availability of these data, which were used under approved permission for the current study, and thus are not publicly available but are available from ICBM upon request. Source data are provided with this paper.


