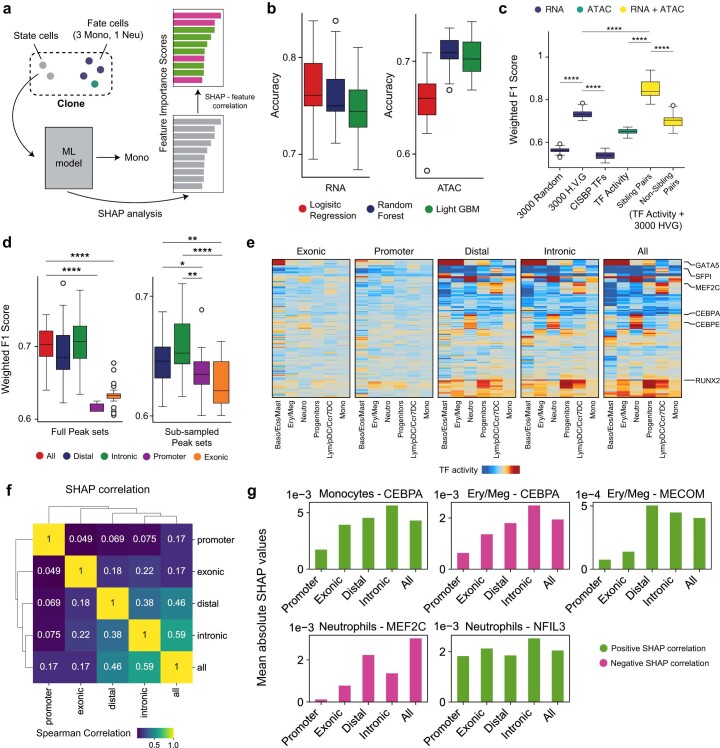
Extended Data Fig. 5. Machine learning analysis to predict cell fate from state.
(a) Schematic of state-fate prediction analysis. (b) Accuracy values obtained with the three model architectures for either RNA (left) or ATAC (right) data (n=25 accuracy values/boxplot). (c) Same plot as Fig. 2m but for F1-weighted scores (Mann Whitney Wilcoxon test, two-sided, n=25 values/boxplot). (d) Boxplots showing variation in F1-weighted score values for ATAC models trained on full peak sets for ‘all’, ‘distal’, ‘intronic’, ‘exonic’ or ‘promoter’ peaks (left) and subsetted ‘distal’, ‘intronic’, ‘exonic’ and ‘promoter’ peak sets(right; n = 8823 peaks; Mann Whitney Wilcoxon test, two-sided; n=25 accuracy values/boxplot). (e) Heatmaps depicting mean TF activity scores for fate predictive TFs across groups of state siblings. TFs show strong fate biased enrichment patterns in ‘distal’, ‘intronic’ and ‘all’ peaks but not exonic and promoter datasets. (f) Heatmap depicting Rank correlation of SHAP values for top predictive TFs shows high similarity between ‘distal’, ‘intronic’ and ‘all’ peaks models. (g) Bar plots of mean absolute SHAP values for a few TFs for fates as indicated. Bars are colored based on value of SHAP correlation. SHAP analysis reveals that motif activity of many lineage specifying TFs is less predictive of cell fate in ‘promoter’ and ‘exonic’ models, while remains comparable across models for some others. Positive SHAP correlation for a feature in a given fate implies that higher values of the feature lead to higher probability of the model outputting that fate label. Negative correlation indicates lower values of the feature lead to higher probability of the model outputting that fate label. All boxplots: center line, median; box limits, first and third quartiles; whiskers, 1.5x interquartile range. For c and d: p-values: **** = p < 0.0001; ** = p < 0.01; * = p < 0.05. Exact p-values in Supplementary Table 12.