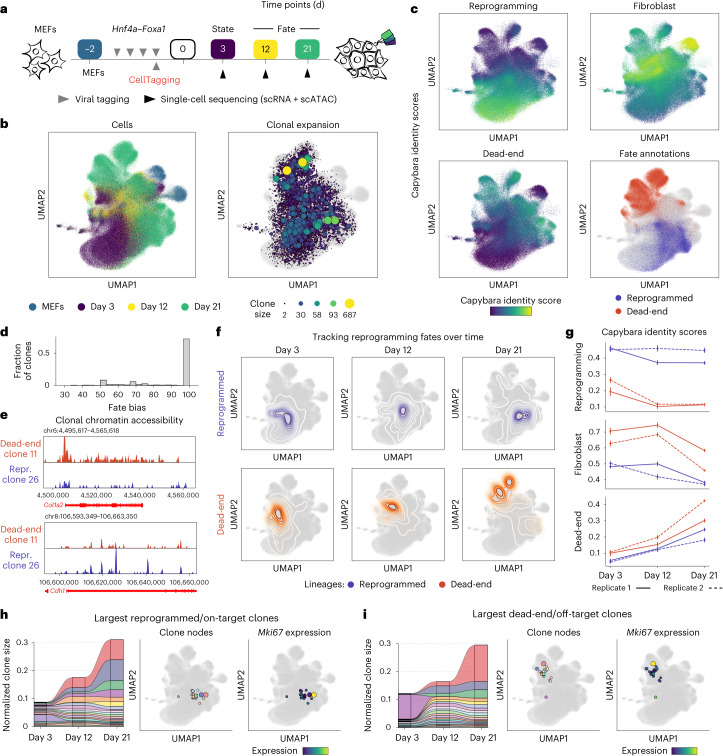
Fig. 3. Application of CellTag-multi to dissect clonal fate dynamics in direct reprogramming.
a, Experimental design for the direct reprogramming state–fate experiment. b, Cells from both scRNA-seq and scATAC-seq, across all time points, were co-embedded with clones and visualized using a UMAP. Left, time point information projected on cells. Right, clonal expansion visualized using clone nodes. c, Capybara transcriptional identity scores projected on scRNA-seq cells for reprogrammed, dead-end and fibroblast cell identities, based on a previous lineage-tracing dataset7. Cell fates were annotated for days 12 and 21. Reprogrammed and dead-end cell fates are highlighted (lower right). d, Histogram of fate bias scores across all state–fate clones. Fate bias scores were calculated using cells from days 12 and 21. e, Clonal chromatin accessibility browser tracks for one dead-end and one reprogramming clone. f, Contour plots showing longitudinal tracking of cell fates enabled by CellTag-multi. g, Transcriptional identity dynamics tracked along both lineages. Dead-end cells depart from a MEF-like identity and acquire an off-target reprogrammed state. h,i, Significant clonal expansion is observed along both lineages, as depicted via alluvial plots, clone nodes and clonal expression levels of Mki67 (a proliferation marker gene) in the 20 largest (h) reprogramming/on-target and (i) dead-end/off-target clones.