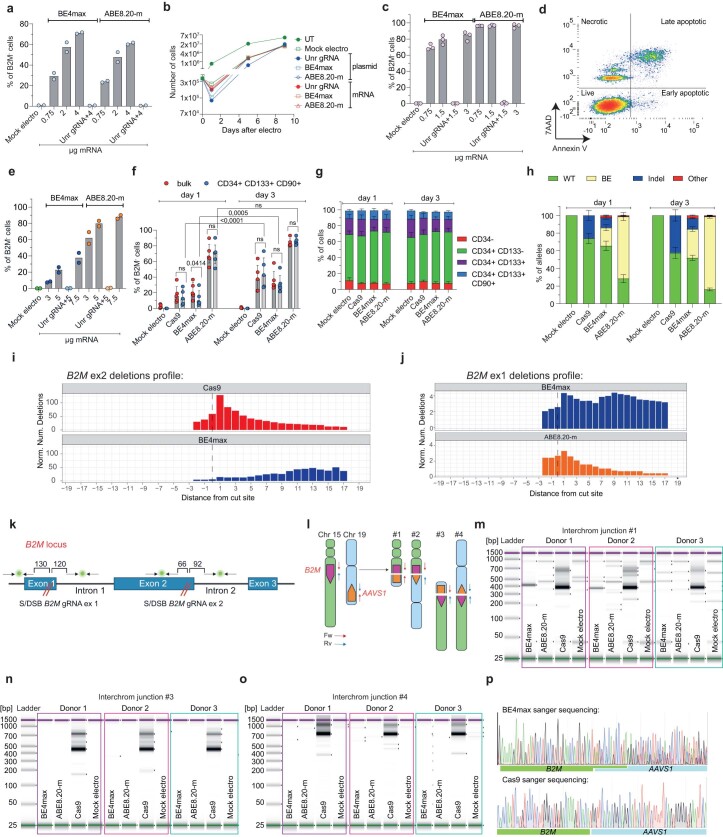
Extended Data Fig. 1. Efficient base editing in human HSPCs and repair outcomes at the target site.
a, Percentage of B2M− B-lymphoblastoid cells measured by flow cytometry 7 days after editing with different mRNA doses (n = 2). Unr: unrelated. Median. b, Growth curve of B-lymphoblastoid cells after treatments (n = 2). Median. c, Percentage of B2M− T cells after editing with different mRNA doses (n = 3). Median. d, Representative plot showing gating strategy for live, early/late apoptotic and necrotic T cells. e, Percentage of B2M− CB HSPCs after editing with different mRNA doses (n = 2). Median. f, Percentage of B2M− mPB HSPCs edited at day 1 or day 3 post-thawing (n = 5). Median with IQR. LME followed by post hoc analysis. g, Proportion of cellular subpopulations within mPB HSPCs from experiments in ‘f’ (n = 5). Mean s.e.m. LME followed by post hoc analysis. h, Percentage of B2M alleles, measured by deep sequencing analysis, being WT or carrying the described editing outcomes in mPB HSPCs (n = 4,5,5,5 for day 1; n = 3,4,4,4 for day 3). Mean s.e.m. i, Distribution of the distance of indels from the B2M exon 2 S/DSB cut site in Cas9-edited (top) and BE4max-edited samples (bottom). j, Distribution of the distance of indels from the B2M exon 1 S/DSB cut site in BE4max-edited (top) and ABE8.20-m-edited samples (bottom). k, Schematic representation of the probes used for deletions detection at B2M target sites in exon 2 and exon 1. The distances between the target site and the closest primer of the ddPCR amplicons are shown. l, Schematic representation of translocations expected upon multiplexed B2M and AAVS1 targeting. m-o, Representative capillary electropherogram showing amplification of #1 (m), #3 (n) and #4- (o) interchromosomal junctions upon HSPC editing with two gRNAs targeting B2M exon 2 and AAVS1 (n = 3). p, Representative Sanger sequencing plot of B2M exon 2-AAVS1 junction in samples from Fig. 1s. All statistical tests are two-tailed. n indicate biologically independent experiments.