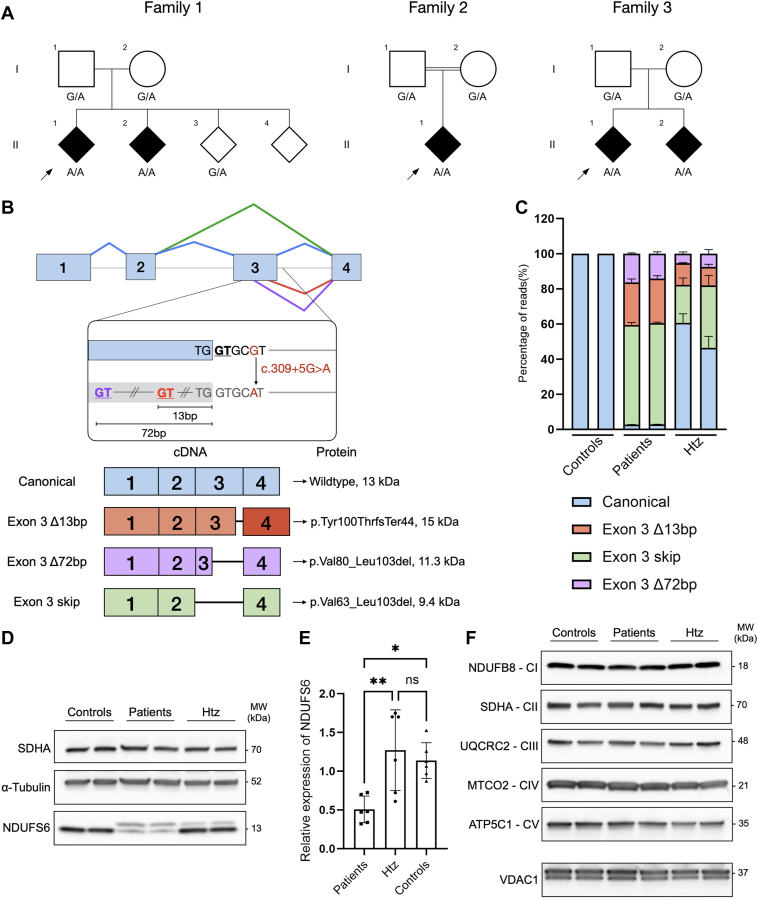
Figure 1.
Genetic and functional analysis of the NDUFS6 c.309+5G>A splice variant. A. Segregation analysis of NDUFS6 c.309+5G>A in families 1-3. Black symbols indicate affected individuals. A double line indicates reported consanguinity. Index patients are indicated with a black arrowhead. B. Top panel: NDUFS6 (NM_004553.6) is schematically shown with exons 1-4 (blue rectangles) and introns (gray line). Caret-like exon connecting lines depict canonical (blue), exon 3 skip (green), exon 3 Δ72bp (purple), and exon 3 Δ13bp (red) splicing events. The 3’-end of exon 3 and intron 3 are presented in more detail to indicate the location of the NDUFS6 c.309+5G>A splice variant (red), the canonical splice-donor site (black and underlined) and the cryptic splice-donor sites (red and purple underlined) located at 13 bp and 72 bp upstream of the 3’-end of exon 3, respectively. Bottom panel: schematic representation of the NDUFS6 transcripts detected in the patients and parents from family 1. The identity of each transcript is indicated on the left, the impact of each splicing on the protein is indicated on the right. C. The relative quantification of NDUFS6 transcripts sequenced by cDNA T-LRS among the 3 genotypes of interest (heterozygotes, patients, and controls). The chart represents the percentage of reads for each transcript quantified relative to the total amount of reads per sample. The bars indicate the standard error of the mean (n = 2). D. Western blot analysis of total protein lysates isolated from lymphoblasts of heterozygotes, patients, and controls with a monoclonal rabbit anti-human NDUFS6 antibody. Monoclonal mouse anti-SDHA and anti-α-tubulin were used as controls for equal loading of mitochondrial and cytosolic fractions, respectively. E. Relative quantification of different NDUFS6 isoforms present in lymphoblasts of heterozygotes, patients, and controls. NDUFS6 band intensity is relative to α-tubulin and normalized to controls. Bar charts are represented as the standard error of the mean (n = 3 for each genotype, with 2 biological replicates for each genotype). F. Immunoblotting of mitochondrial fractions from 3 genotypes of interest (heterozygotes, patients, and controls) with monoclonal mouse anti-human NDUFB8, SDHA, UQCR2, and ATP5C1 and monoclonal rabbit anti-human MTCO2 antibodies to target 1 subunit from each OXPHOS complex. Monoclonal mouse anti-human VDAC1 was used as a control for mitochondrial proteins. All statistical analyses were performed using one-way ANOVA. Asterisks denote significance after Tukey’s multiple comparison correction. Abbreviations: Htz, heterozygotes; ∗∗P < .01, ∗P < .05; ns, not significant; SDHA, succinate dehydrogenase complex subunit A.