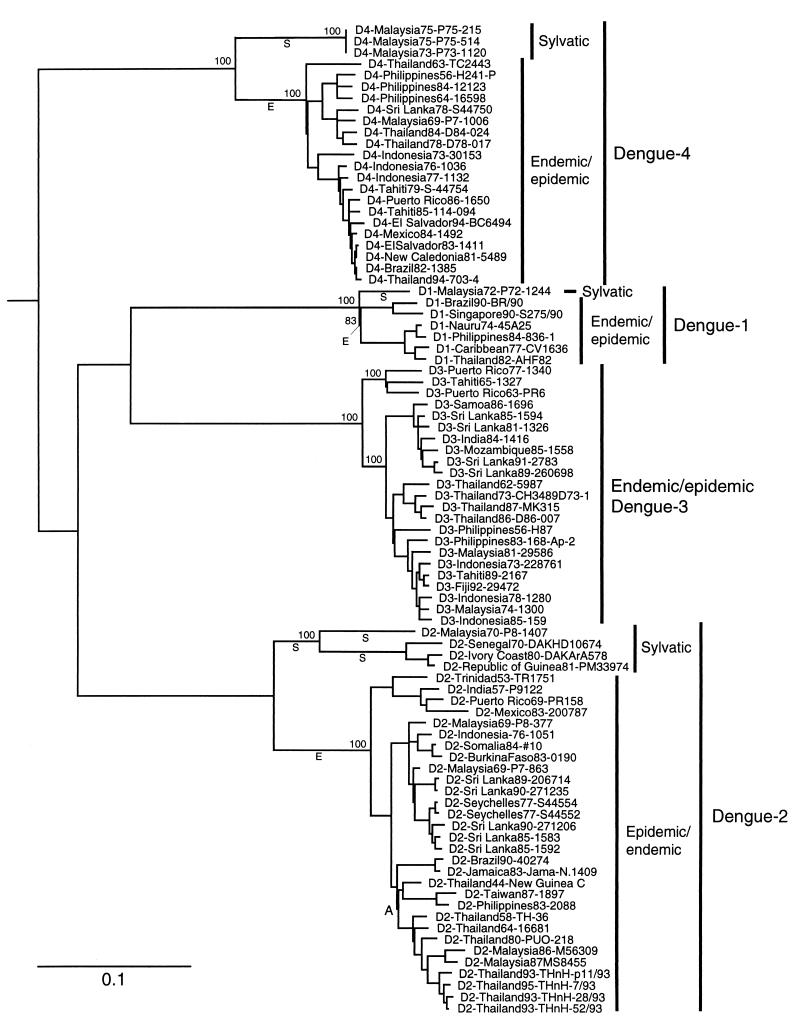
FIG. 1.
Phylogenetic tree derived from E protein gene nucleotide sequences of sylvatic and representative endemic/epidemic DEN strains using maximum parsimony (PAUP, version 3.0) and drawn using branch lengths derived by neighbor joining using the Kimura two-parameter formula (PHYLIP, version 3.5p). The scale shows a genetic distance of 0.1, or 10% nucleotide sequence divergence. Homologous sequences from Japanese encephalitis, Kunjin, and West Nile viruses (a sister group to DEN [20]) were used as the outgroup to root the DEN tree. Numbers indicate bootstrap (8) values for monophyletic groups to the right. Branches labeled “E” indicate periods of evolution predicted to represent emergence of endemic/epidemic DEN and used to generate Table 3; branches labeled “S” represent continued sylvatic evolution during the same time period. Node A defines the clade used for the regression analysis of the DEN evolutionary rate.