Figure 1.
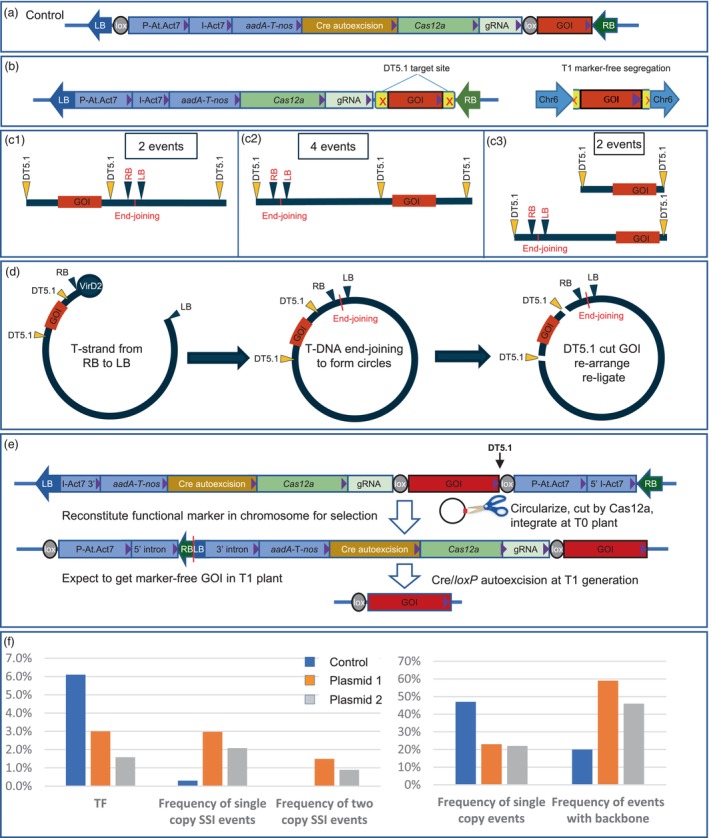
T‐DNA structures in Agrobacterium binary vectors and transgenic soybean plants and the frequencies of transgenic events. (a) T‐DNA configuration from the control binary vector with intact marker gene. (b) T‐DNA structure with two DT5.1 target sites flanking the GOI (left) and the expected GOI integration at the DT5.1 site (right) in soybean chromosome 6 (Chr6) after Cas12a released the GOI from the T‐DNA. (c) Sequence analysis of the eight single copy target events revealed RB‐LB junctions with three different re‐arrangements at the DT5.1 site. (d) Proposed T‐circle formation before integration. Left: T‐strand from T‐DNA in (b) initiated from RB and stopped at LB; middle: converted to T‐DNA, RB‐LB end joining to form T‐circle; right: DT5.1 sites in the vector were cut, re‐arrange, re‐ligate at one end and insert into the chromosomal DT5.1. (e) The T‐circle vector design to reduce random insertion events and enrich target events. (f) Summarized frequencies of transformation, SSI events, single copy events and the events with backbone. P‐At.Act7, Arabidopsis Actin 7 promoter; I‐Act7, P‐At.Act7 intron; aadA, Encoding aminoglycoside‐3″‐adenylyltransferase conferring spectinomycin resistance; T‐nos, Nopaline synthase terminator; Cre, Cre autoexcision cassette; Cas12a and gRNA, LbCas12a And guide RNA expression cassettes, respectively; I‐Act7 3′ and I‐Act7 5′, 270 bp of Actin 7 intron 3′ and 5′ sequence, respectively.
