Abstract
Increasing evidence shows that flaws in machine learning (ML) algorithm validation are an underestimated global problem. In biomedical image analysis, chosen performance metrics often do not reflect the domain interest, and thus fail to adequately measure scientific progress and hinder translation of ML techniques into practice. To overcome this, we created Metrics Reloaded, a comprehensive framework guiding researchers in the problem-aware selection of metrics. Developed by a large international consortium in a multi-stage Delphi process, it is based on the novel concept of a problem fingerprint – a structured representation of the given problem that captures all aspects that are relevant for metric selection, from the domain interest to the properties of the target structure(s), data set and algorithm output. Based on the problem fingerprint, users are guided through the process of choosing and applying appropriate validation metrics while being made aware of potential pitfalls. Metrics Reloaded targets image analysis problems that can be interpreted as classification tasks at image, object or pixel level, namely image-level classification, object detection, semantic segmentation, and instance segmentation tasks. To improve the user experience, we implemented the framework in the Metrics Reloaded online tool. Following the convergence of ML methodology across application domains, Metrics Reloaded fosters the convergence of validation methodology. Its applicability is demonstrated for various biomedical use cases.
Introduction
Automatic image processing with machine learning (ML) is gaining increasing traction in biological and medical imaging research and practice. Research has predominantly focused on the development of new image processing algorithms. The critical issue of reliable and objective performance assessment of these algorithms, however, remains largely unexplored. Algorithm performance in image processing is commonly assessed with validation metrics1 that should serve as proxies for the domain interest. In consequence, the impact of validation metrics cannot be overstated; first, they are the basis for deciding on the practical (e.g. clinical) suitability of a method and are thus a key component for translation into biomedical practice. In fact, validation that is not conducted according to relevant metrics could be one major reason for why many artificial intelligence (AI) developments in medical imaging fail to reach clinical practice [32, 72]. In other words, the numbers presented in journals and conference proceedings do not reflect how successful a system will be when applied in practice. Second, metrics guide the scientific progress in the field; flawed metric use can lead to entirely futile resource investment and infeasible research directions while obscuring true scientific advancements.
Despite the importance of metrics, an increasing body of work shows that the metrics used in common practice often do not adequately reflect the underlying biomedical problems, diminishing the validity of the investigated methods [16, 23, 26, 35, 37, 47, 51, 82, 85]. This especially holds true for challenges, internationally respected competitions that have become the de facto standard for comparative performance assessment of image processing methods. These challenges are often published in prestigious journals [11, 69, 83] and receive tremendous attention from both the scientific community and industry. Among a number of shortcomings in design and quality control that were recently unveiled by a multi-center initiative [47], the choice of inappropriate metrics stood out as a core problem. Compared to other areas of AI research, choosing the right metric is particularly challenging in image processing because the suitability of a metric depends on various factors. As a foundation for the present work, we identified three core categories related to pitfalls in metric selection (see Fig. 1a):
Inappropriate choice of the problem category: The chosen metrics do not always reflect the biomedical need. For example, object detection problems are often framed as segmentation tasks, resulting in the use of metrics that do not account for the potentially critical localization of all objects in the scene [9, 29] (Fig. 1a, top left).
Poor metric selection: Certain characteristics of a given biomedical problem render particular metrics inadequate. Mathematical metric properties are often neglected, for example, when using the Dice Similarity Coefficient (DSC) in the presence of particularly small structures (Fig. 1a, top right).
Poor metric application: Even if a metric is well-suited for a given problem in principle, pitfalls can occur when applying that metric to a specific data set. For example, a common flaw pertains to ignoring hierarchical data structure, as in data from multiple hospitals or a variable number of images per patient (Fig. 1a, bottom), when aggregating metric values.
Figure 1: Contributions of the Metrics Reloaded framework.
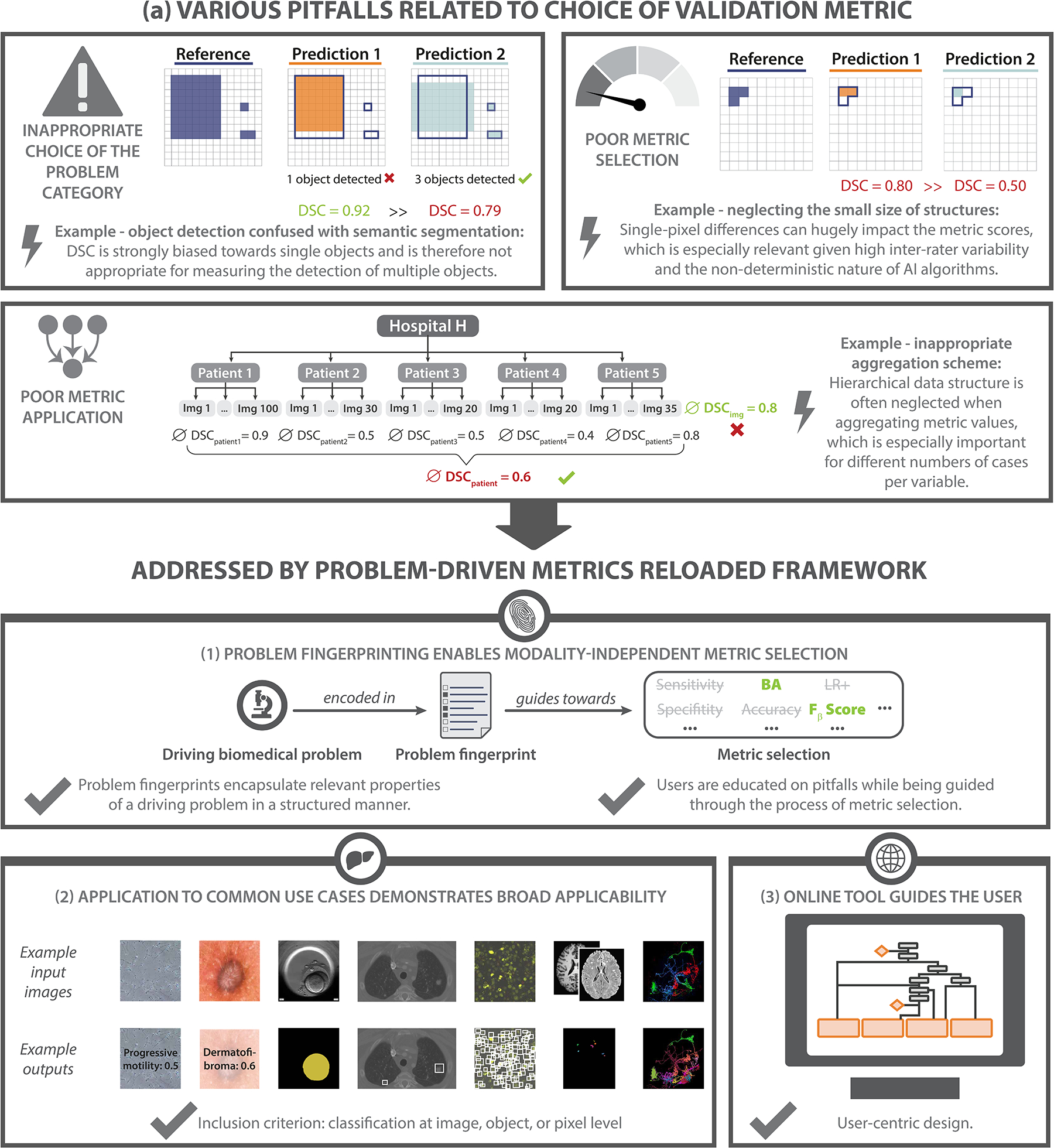
a) Motivation: Common problems related to metrics typically arise from (top left) inappropriate choice of the problem category (here: object detection confused with semantic segmentation), (top right) poor metric selection (here: neglecting the small size of structures) and (bottom) poor metric application (here: inappropriate aggregation scheme). Pitfalls are highlighted by lightning bolts, ∅ refers to the average Dice Similarity Coefficient (DSC) values. Green metric values correspond to a good metric value, whereas red values correspond to a poor value. Green check marks indicate desirable behavior of metrics, red crosses indicate undesirable behavior. b) Metrics Reloaded addresses these pitfalls. (1) To enable the selection of metrics that match the domain interest, the framework is based on the new concept of problem fingerprinting, i.e., the generation of a structured representation of the given biomedical problem that captures all properties that are relevant for metric selection. Based on the problem fingerprint, Metrics Reloaded guides the user through the process of metric selection and application while raising awareness of relevant pitfalls. (2) An instantiation of the framework for common biomedical use cases demonstrates its broad applicability. (3) A publicly available online tool facilitates application of the framework.
These problems are magnified by the fact that common practice often grows historically, and poor standards may be propagated between generations of scientists and in prominent publications. To dismantle such historically grown poor practices and leverage distributed knowledge from various subfields of image processing, we established the multidisciplinary Metrics Reloaded2 consortium. This consortium comprises international experts from the fields of medical image analysis, biological image analysis, medical guideline development, general ML, different medical disciplines, statistics and epidemiology, representing a large number of biomedical imaging initiatives and societies.
The mission of Metrics Reloaded is to foster reliable algorithm validation through problem-aware, standardized choice of metrics with the long-term goal of (1) enabling the reliable tracking of scientific progress and (2) aiding to bridge the current chasm between ML research and translation into biomedical imaging practice.
Based on a kickoff workshop held in December 2020, the Metrics Reloaded framework (Fig. 1b and Fig. 2) was developed using a multi-stage Delphi process [7] for consensus building. Its primary purpose is to enable users to make educated decisions on which metrics to choose for a driving biomedical problem. The foundation of the metric selection process is the new concept of problem fingerprinting (Fig. 3). Abstracting from a specific domain, problem fingerprinting is the generation of a structured representation of the given biomedical problem that captures all properties relevant for metric selection. As depicted in Fig. 3, the properties captured by the fingerprint comprise domain interest-related properties, such as the particular importance of structure boundary, volume or center, target structure-related properties, such as the shape complexity or the size of structures relative to the image grid size, data set-related properties, such as class imbalance, as well as algorithm output-related properties, such as the theoretical possibility of the algorithm output not containing any target structure.
Figure 2: Metrics Reloaded recommendation framework from a user perspective.
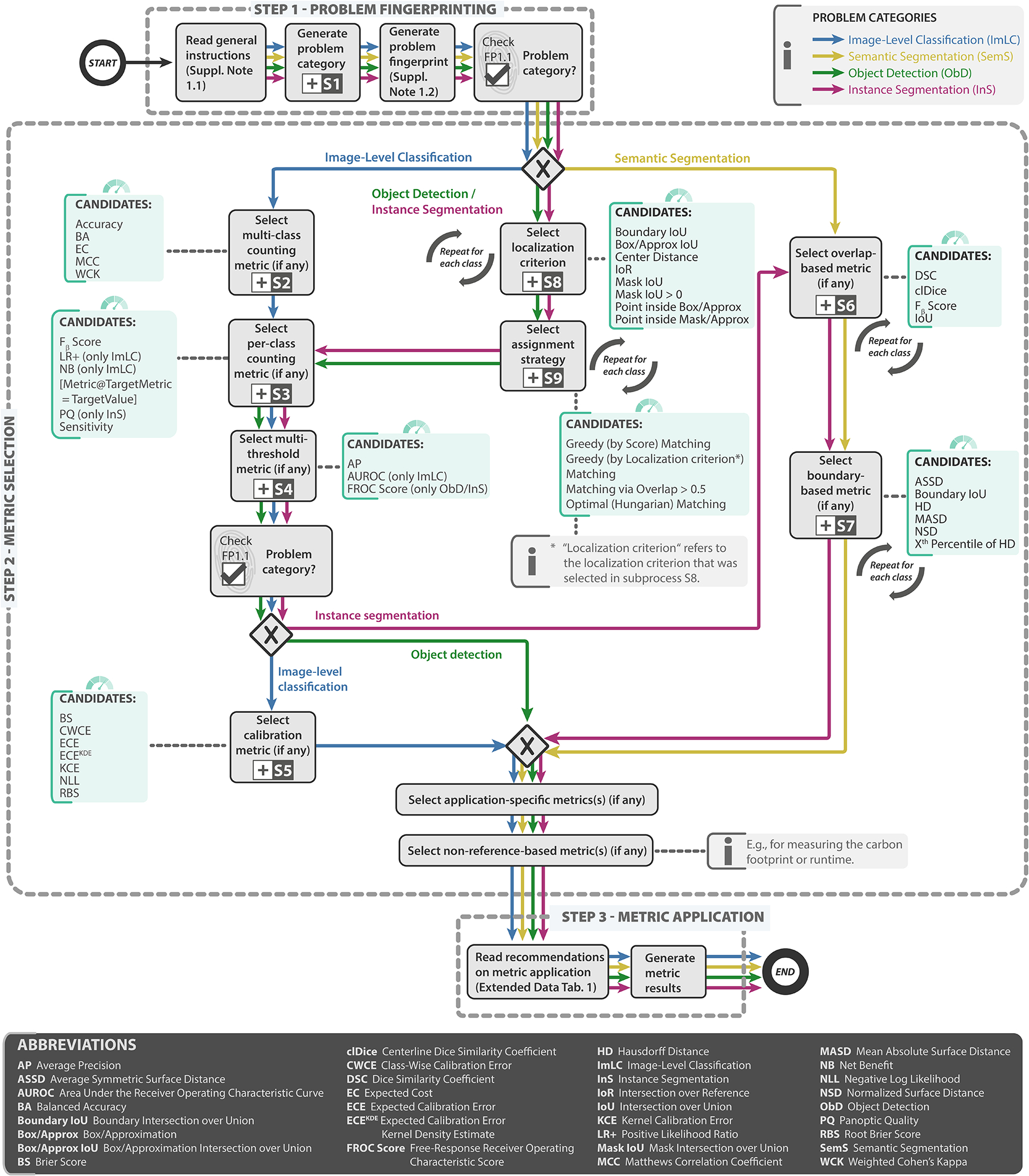
In step 1 - problem fingerprinting, the given biomedical image analysis problem is mapped to the appropriate image problem category, namely image-level classification (ImLC), semantic segmentation (SemS), object detection (ObD), or instance segmentation (InS) (Fig. 4). The problem category and further characteristics of the given biomedical problem relevant for metric selection are then captured in a problem fingerprint (Fig. 3). In step 2 - metric selection, the user follows the respective coloured path of the chosen problem category (ImLC →, SemS →, ObD →, or InS →) to select a suitable pool of metrics from the Metrics Reloaded pools shown in green. When a tree branches, the fingerprint items determine which exact path to take. Finally, in step 3 - metric application, the user is supported in applying the metrics to a given data set. During the traversal of the decision tree, the user goes through subprocesses, indicated by the ⊞-symbol, which are provided in Extended Data Figs. 1–9 and represent relevant steps in the metric selection process. Ambiguities related to metric selection are resolved via decision guides (Suppl. Note 2.7) that help users make an educated decision when multiple options are possible. A comprehensive textual description of the recommendations for all four problem categories as well as for the selection of corresponding calibration metrics (if any) is provided in Suppl. Note 2.2 - Suppl. Note 2.6. An overview of the symbols used in the process diagram is provided in Fig. SN 5.1. Condensed versions of the mappings for every category can be found in Suppl. Note 2.2 for image-level classification, Suppl. Note 2.3 for semantic segmentation, Suppl. Note 2.4 for object detection, and Suppl. Note 2.5 for instance segmentation.
Figure 3: Relevant properties of a driving biomedical image analysis problem are captured by the problem fingerprint.
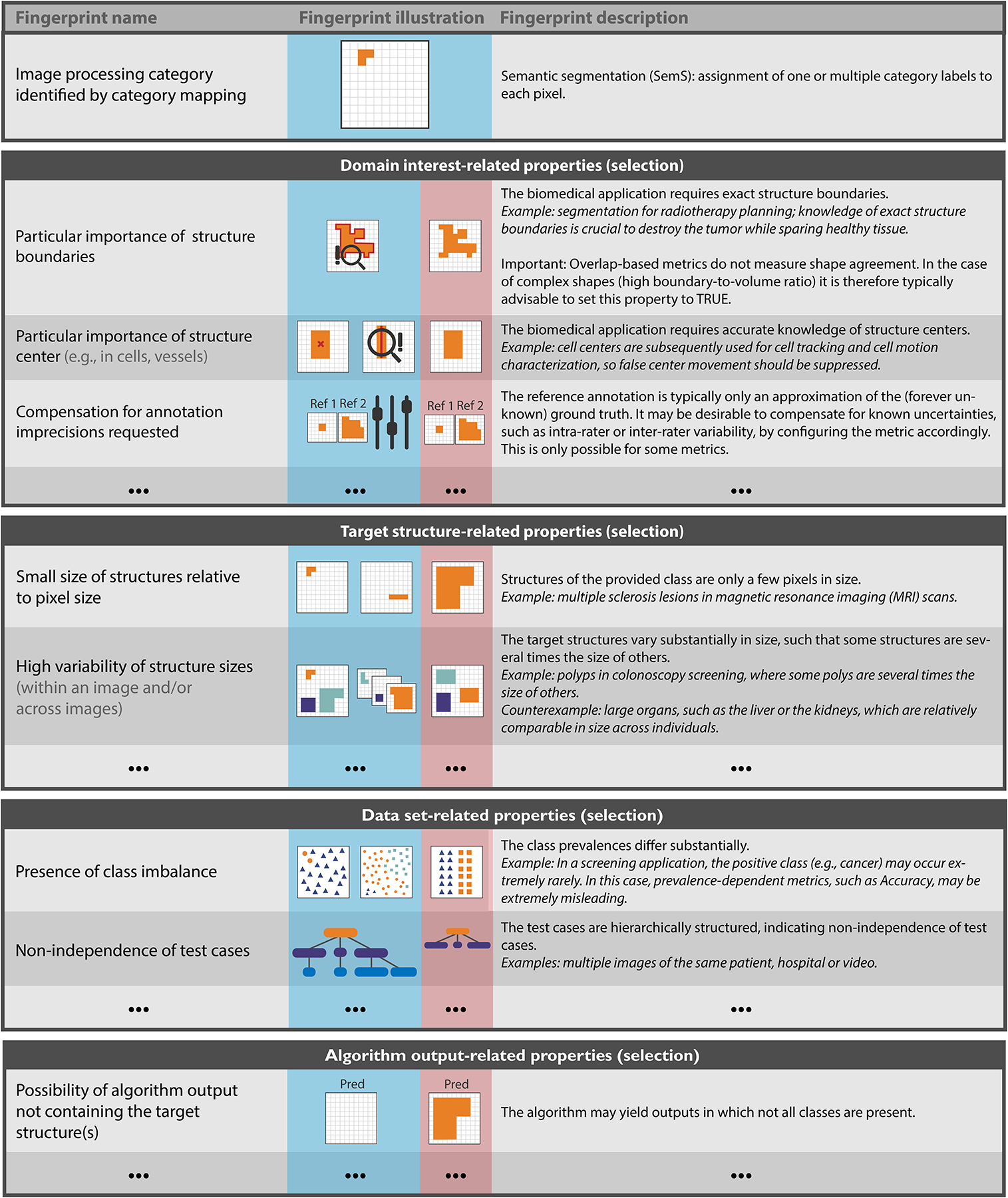
(selection for semantic segmentation shown here). The fingerprint comprises a set of items, each of which represents a specific property of the problem, is either binary or categorical, and must be instantiated by the user. Besides the problem category, the fingerprint comprises domain interest-related, target structure-related, data set-related and algorithm output-related properties. A comprehensive version of the fingerprints for all problem categories can be found in Figs. SN 2.7–SN 2.9 (image-level classification), Figs. SN 2.10/SN 2.11 (semantic segmentation), Figs. SN 2.12–SN 2.14 (object detection) and Figs. SN 2.15–SN 2.17 (instance segmentation). Used abbreviations: Prediction (Pred), Reference (Ref).
Based on the problem fingerprint, the user is then, in a transparent and understandable manner, guided through the process of selecting an appropriate set of metrics while being made aware of potential pitfalls related to the specific characteristics of the underlying biomedical problem. The Metrics Reloaded framework currently supports problems in which categorical target variables are to be predicted based on a given n-dimensional input image (possibly enhanced with context information) at pixel, object or image level, as illustrated in Fig. 4. It thus supports problems that can be assigned to one of the following four problem categories: image-level classification (image level), object detection (object level), semantic segmentation (pixel level), or instance segmentation (pixel level). Designed to be imaging modality-independent, Metrics Reloaded can be suited for application in various image analysis domains even beyond the field of biomedicine.
Figure 4: Metrics Reloaded fosters the convergence of validation methodology across modalities, application domains and classification scales.
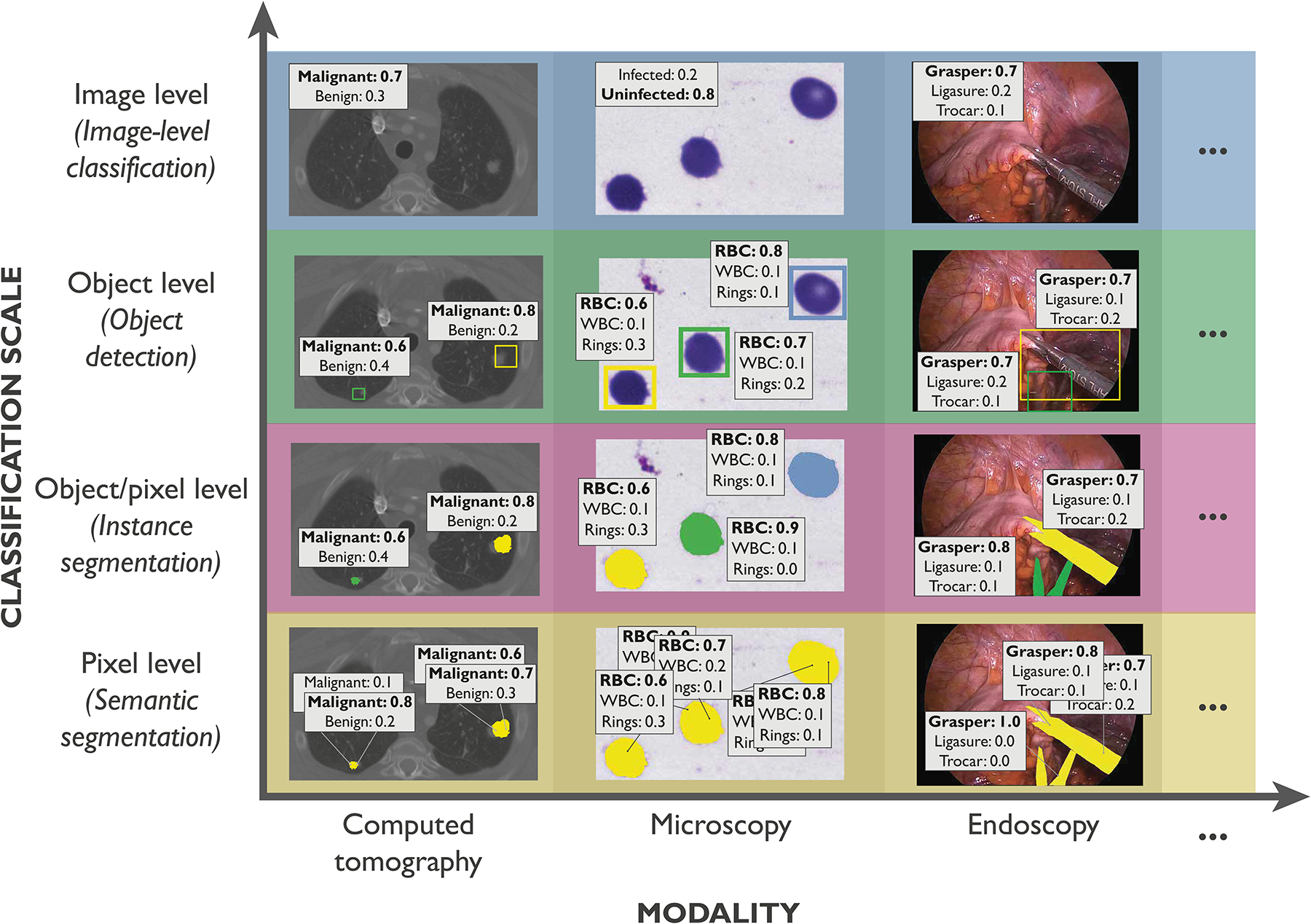
The framework considers problems in which categorical target variables are to be predicted at image, object and/or pixel level, resulting (from top to bottom) in image-level classification, object detection, instance segmentation or semantic segmentation problems. These problem categories are relevant across modalities (here computed tomography (CT), microscopy and endoscopy) and application domains. From left to right: annotation of (left) benign and malignant lesions in CT images [3], (middle) different cell types in microscopy images [46], and (right) medical instruments in laparoscopy images [48].
Here, we present the key contributions of our work in detail, namely (1) the Metrics Reloaded framework for problem-aware metric selection along with the key findings and design decisions that guided its development (Fig. 2), (2) the application of the framework to common biomedical use cases, showcasing its broad applicability (selection shown in Fig. 5) and (3) the open online tool that has been implemented to improve the user experience with our framework.
Figure 5: Instantiation of the framework with recommendations for concrete biomedical questions.
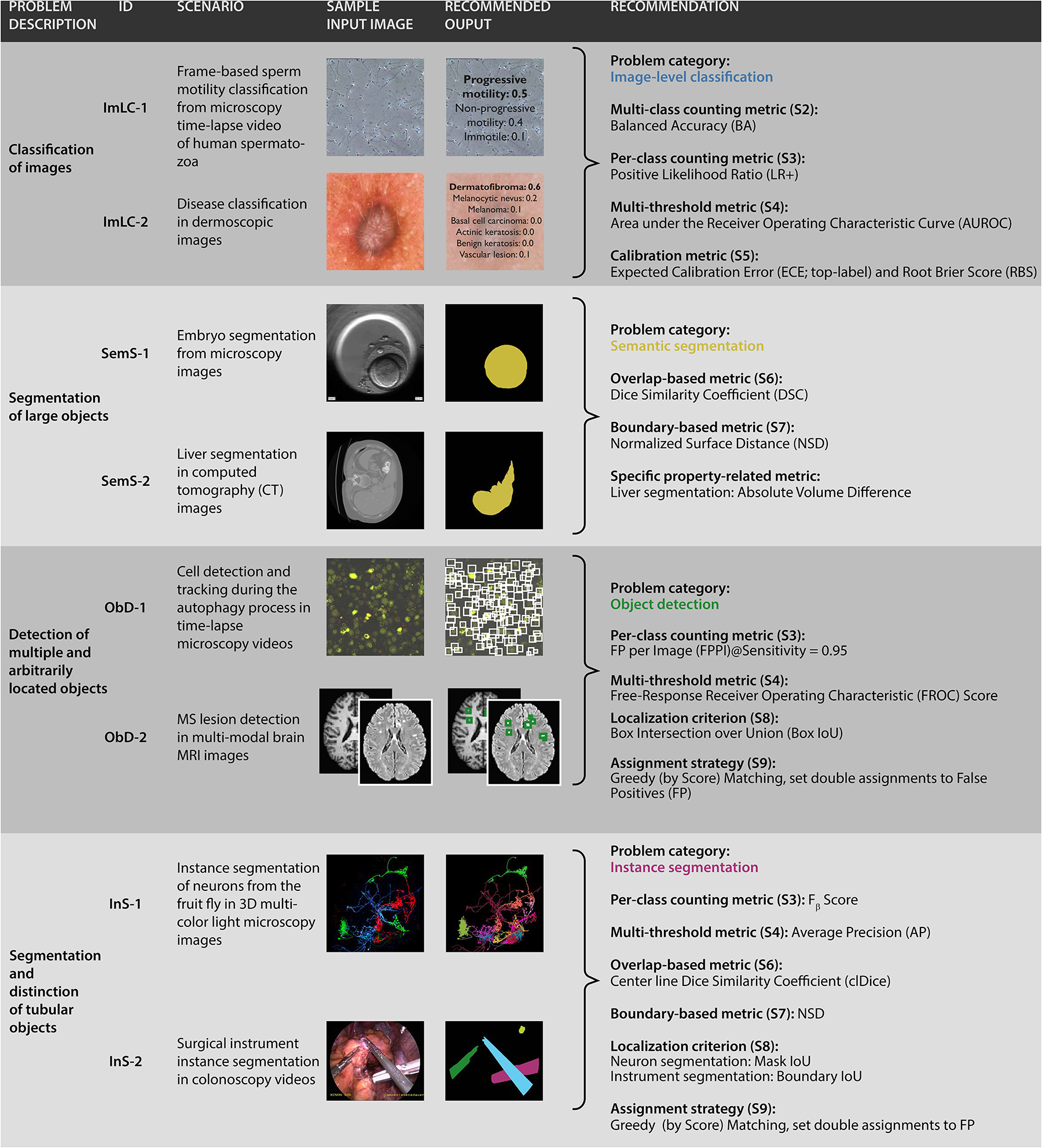
From top to bottom: (1) Image classification for the examples of sperm motility classification [25] and disease classification in dermoscopic images [12, 84]. (2) Semantic segmentation of large objects for the examples of embryo segmentation from microscopy [79] and liver segmentation in computed tomography (CT) images [2, 73]. (3) Detection of multiple and arbitrarily located objects for the examples of cell detection and tracking during the autophagy process [56, 90] and multiple sclerosis (MS) lesion detection in multi-modal brain magnetic resonance imaging (MRI) images [14, 36]. (4) Instance segmentation of tubular objects for the examples of instance segmentation of neurons from the fruit fly [50, 54, 81] and surgical instrument instance segmentation [48]. The corresponding traversals through the decision trees are shown in Suppl. Note 4. An overview of the recommended metrics can be found in Suppl. Note 3.1, including relevant information for each metric.
Metrics Reloaded Framework
Metrics Reloaded is the result of a multi-stage Delphi process, comprising five international workshops, nine surveys, numerous expert group meetings, and crowdsourced feedback processes, all conducted between 2020 and 2022. As a foundation of the recommendation framework, we identified common and rare pitfalls related to metrics in the field of biomedical image analysis using a community-powered process, detailed in this work’s sister publication [65]. We found that common practice is often not well-justified, and poor practices may even be propagated from one generation of scientists to the next. Importantly, many pitfalls generalize not only across the four problem categories that our framework addresses but also across domains (Fig. 4). This is because the source of the pitfall, such as class imbalance, uncertainties in the reference, or poor image resolution, can occur irrespective of a specific modality or application.
Following the convergence of AI methodology across domains and problem categories, we therefore argue for the analogous convergence of validation methodology.
Cross-domain approach enables integration of distributed knowledge
To break historically grown poor practices, we followed a multidisciplinary cross-domain approach that enabled us to critically question common practice in different communities and integrate distributed knowledge in one common framework. To this end, we formed an international multidisciplinary consortium of 73 experts from various biomedical image analysis-related fields. Furthermore, we crowdsourced metric pitfalls and feedback on our approach in a social media campaign. Ultimately, a total of 156 researchers contributed to this work, including 84 mentioned in the acknowledgements. Consideration of the different knowledge and perspectives on metrics led to the following key design decisions for Metrics Reloaded:
Encapsulating domain knowledge: The questions asked to select a suitable metric are mostly similar regardless of image modality or application: Are the classes balanced? Is there a specific preference for the positive or negative class? What is the accuracy of the reference annotation? Is the structure boundary or volume of relevance for the target application? Importantly, while answering these questions requires domain expertise, the consequences in terms of metric selection can largely be regarded as domain-independent. Our approach is thus to abstract from the specific image modality and domain of a given problem by capturing the properties relevant for metric selection in a problem fingerprint (Fig. 3).
Exploiting synergies across classification scales: Similar considerations apply with regard to metric choice for classification, detection and segmentation tasks, as they can all be regarded as classification tasks at different scales (Fig. 4). The similarities between the categories, however, can also lead to problems when the wrong category is chosen (see Fig. 1a, top left). Therefore, we (1) address all four problem categories in one common framework (Fig. 2) and (2) cover the selection of the problem category itself in our framework (Extended Data Fig. 1).
Setting new standards: As the development and implementation of recommendations that go beyond the state of the art often requires critical mass, we involved stakeholders of various communities and societies in our consortium. Notably, out crowdsourcing-based approach led to a pool of metric candidates (Tab. SN 2.1) that not only includes commonly applied metrics, but also metrics that have to date received little attention in biomedical image analysis.
Abstracting from inference methodology: Metrics should be chosen based solely on the driving biomedical problem and not be affected by algorithm design choices. For example, the error functions applied in common neural network architectures do not justify the use of corresponding metrics (e.g. validating with DSC to match the Dice loss used for training a neural network). Instead, the domain interest should guide the choice of metric, which, in turn, can guide the choice of the loss term.
Exploiting complementary metric strengths: A single metric typically cannot cover the complex requirements of the driving biomedical problem [64]. To account for the complementary strengths and weakness of metrics, we generally recommend the usage of multiple complementary metrics to validate image analysis problems. As detailed in our recommendations (Suppl. Note 2), we specifically recommend the selection of metrics from different families.
Validation by consensus building and community feedback: A major challenge for research on metrics is its validation, due to the lack of methods capable of quantitatively assessing the superiority of a given metric set over another. Following the spirit of large consortia formed to develop reporting guidelines (e.g., CONSORT [71], TRIPOD [55], STARD [6]), we built the validation of our framework on three main pillars: (1) Delphi processes to challenge and refine the proposals of the expert groups that worked on individual components of the framework, (2) community feedback obtained by broadcasting the framework via society mailing lists and social media platforms and (3) and instantiation of the framework to a range of different biological and medical use cases.
Involving and educating users: Choosing adequate validation metrics is a complex process. Rather than providing a black box recommendation, Metrics Reloaded guides the user through the process of metric selection while raising awareness on pitfalls that may occur. In cases in which the tradeoffs between different choices must be considered, decision guides (Suppl. Note 2.7) assist in deciding between competing metrics while respecting individual preferences.
Problem fingerprints encapsulate relevant domain knowledge
To encapsulate relevant domain knowledge in a common format and then enable a modality-agnostic metric recommendation approach that generalizes over domains, we developed the concept of problem fingerprinting, illustrated in Fig. 3. As a foundation, we crowdsourced all properties of a driving biomedical problem that are potentially relevant for metric selection via surveys issued to the consortium (see Suppl. Methods). This process resulted in a list of binary and categorical variables (fingerprint items) that must be instantiated by a user to trigger the Metrics Reloaded recommendation process. Common issues often relate to selecting metrics from the wrong problem category, as illustrated in Fig. 1a (top left). To avoid such issues, problem fingerprinting begins with mapping a given problem with all its intrinsic and data set-related properties to the corresponding problem category via the category mapping shown in Extended Data Fig. 1. The problem category is a fingerprint item itself.
In the following, we will refer to all fingerprint items with the notation FPX.Y, where Y is a numerical identifier and the index X represents one of the following families:
FP1 - Problem category refers to the problem category generated by S1 (Extended Data Fig. 1). FP2 - Domain interest-related properties reflect user preferences and are highly dependent on the target application. A semantic image segmentation that serves as the foundation for radiotherapy planning, for example, would require exact contours (FP2.1 Particular importance of structure boundaries = TRUE). On the other hand, for a cell segmentation problem that serves as prerequisite for cell tracking, the object centers may be much more important (FP2.3 = TRUE). Both problems could be tackled with identical network architectures, but the validation metrics should be different.
FP3 - Target structure-related properties represent inherent properties of target structure(s) (if any), such as the size, size variability and the shape. Here, the term target structures can refer to any object/structure of interest, such as cells, vessels, medical instruments or tumors.
FP4 - Data set-related properties capture properties inherent to the provided data to which the metric is applied. They primarily relate to class prevalences, uncertainties of the reference annotations, and whether the data structure is hierarchical.
FP5 - Algorithm output-related properties encode properties of the output, such as the availability of predicted class scores.
Note that not all properties are relevant for all problem categories. For example, the shape and size of target structures is highly relevant for segmentation problems but irrelevant for image classification problems. The complete problem category-specific fingerprints are provided in Suppl. Note 1.3.
Metrics Reloaded addresses all three types of metric pitfalls
Metrics Reloaded was designed to address all three types of metric pitfalls identified in [65] and illustrated in Fig. 1a. More specifically, each of the three steps shown in Fig. 2 addresses one type of pitfall:
Step 1 - Fingerprinting.
A user should begin by reading the general instructions of the recommendation framework, provided in Suppl. Note 1.1. Next, the user should convert the driving biomedical problem to a problem fingerprint. This step is not only a prerequisite for applying the framework across application domains and classification scales, but also specifically addresses the inappropriate choice of the problem category via the integrated category mapping. Once the user’s domain knowledge has been encapsulated in the problem fingerprint, the actual metric selection is conducted according to a domain- and modality-agnostic process.
Step 2 - Metric Selection.
A Delphi process yielded the Metrics Reloaded pool of reference-based validation metrics shown in Tab. SN 2.1. Notable, this pool contains metrics that are currently not widely known in some biomedical image analysis communities. A prominent example is the Net Benefit (NB) [87] metric, popular in clinical prediction tasks and designed to determine whether basing decisions on a method would do more good than harm. A diagnostic test, for example, may lead to early identification and treatment of a disease, but typically will also cause a number of patients without disease being subjected to unnecessary further interventions. NB allows to consider such tradeoffs by putting benefits and harms on the same scale so that they can be directly compared. Another example is the Expected Cost (EC) metric [42], which can be seen as a generalization of Accuracy with many desirable added features, but is not well-known in the biomedical image analysis communities [21]. Based on the Metrics Reloaded pool, the metric recommendation is performed with a Business Process Model and Notation (BPMN)-inspired flowchart (see Fig. SN 5.1), in which conditional operations are based on one or multiple fingerprint properties (Fig. 2). The main flowchart has three substeps, each addressing the complementary strengths and weaknesses of common metrics. First, common reference-based metrics, which are based on the comparison of the algorithm output to a reference annotation, are selected. Next, the pool of standard metrics can be complemented with custom metrics to address application-specific complementary properties. Finally, non-reference-based metrics assessing speed, memory consumption or carbon footprint, for example, can be added to the metric pool(s). In this paper, we focus on the step of selecting reference-based metrics, because this is where synergies across modalities and scales can be exploited.
These synergies are showcased by the substantial overlap between the different paths that, depending on the problem category, are taken through the mapping during metric selection. All paths comprise several subprocesses S (indicated by the ⊞-symbol), each of which holds a subsidiary decision tree representing one specific step of the selection process. Traversal of a subprocess typically leads to the addition of a metric to the problem-specific metric pool. In multi-class prediction problems, dedicated metric pools for each class may need to be generated as relevant properties may differ from class to class. A three-dimensional semantic segmentation problem, for example, could require the simultaneous segmentation of both tubular and non-tubular structures (e.g., liver vessels and tissue). These require different metrics for validation. Although this is a corner case, our framework addresses this issue in principle. In ambiguous cases, i.e., when the user can choose between two options in one step of the decision tree, a corresponding decision guide details the tradeoffs that need to be considered (Suppl. Note 2.7). For example, the Intersection over Union (IoU) and the DSC are mathematically closely related. The concrete choice typically boils down to a simple user or community preference.
Fig. 2 along with the corresponding Subprocesses S1–S9 (Extended Data Fig. 1–Extended Data Fig. 9) captures the core contribution of this paper, namely the consensus recommendation of the Metrics Reloaded consortium according to the final Delphi process. For all ten components, the required Delphi consensus threshold (>75% agreement) was met. In all cases of disagreement, which ranged from 0% to 7% for Fig. 2 and S1–S9, each remaining point of criticism was respectively only raised by a single person. The following paragraphs present a summary of the four different colored paths through Step 2 - Metric Selection of the recommendation tree (Fig. 2) for the task of selecting reference-based metrics from the Metrics Reloaded pool of common metrics. More comprehensive textual descriptions can be found in Suppl. Note 2.
Image-level Classification (ImLC).
Image-level classification is conceptually the most straight- forward problem category, as the task is simply to assign one of multiple possible labels to an entire image (see Suppl. Note 2.2). The validation metrics are designed to measure two key properties: discrimination and calibration.
Discrimination refers to the ability of a classifier to discriminate between two or more classes. This can be achieved with counting metrics that operate on the cardinalities of a fixed confusion matrix (i.e., the true/false positives/negatives in the binary classification case). Prominent examples are Sensitivity, Specificity or F1 Score for binary settings and Matthews Correlation Coefficient (MCC) for multi-class settings. Converting predicted class scores to a fixed confusion matrix (in the binary case by setting a potentially arbitrary cutoff) can, however, be regarded as problematic in the context of performance assessment [65]. Multi-threshold metrics, such as Area under the Receiver Operating Characteristic Curve (AUROC), are therefore based on varying the cutoff, which enables the explicit analysis of the tradeoff between competing properties such as Sensitivity and Specificity.
While most research in biomedical image analysis focuses on the discrimination capabilities of classifiers, a complementary important property is the calibration of a model. An uncertainty-aware model should yield predicted class scores that represent the true likelihood of events [24], as detailed in Suppl. Note 2.6. Overoptimistic or underoptimistic classifiers can be especially problematic in prediction tasks where a clinical decision may be made based on the risk of the patient of developing a certain condition. Metrics Reloaded hence provides recommendations for validating the algorithm performance both in terms of discrimination and calibration. We recommend the following process for classification problems (blue path in Fig. 2; detailed description in Suppl. Note 2.2):
-
Select multi-class metric (if any): Multi-class metrics have the unique advantage of capturing the performance of an algorithm for all classes in a single value. With the ability of taking into account all entries of the multi-class confusion matrix, they provide a holistic measure of performance without the need for customized class-aggregation schemes. We recommend using a multi-class metric if a decision rule applied to the predicted class scores is available (FP2.6). In certain use cases, especially in the presence of ordinal data, there is an unequal severity of class confusions (FP2.5.2), meaning that different costs should be applied to different misclassifications reflected by the confusion matrix. In such cases, we generally recommend EC as metric. Otherwise, depending on the specific scenario, Accuracy, Balanced Accuracy (BA) and MCC may be viable alternatives. The concrete choice of metric depends primarily on the prevalences (e.g. frequencies) of classes in the provided validation set and the target population (FP4.1/2), as detailed in Subprocess S2 (Extended Data Fig. 2) and the corresponding textual description in Suppl. Note 2.2.
As class-specific analyses are not possible with multi-class metrics, which can potentially hide poor performance on individual classes, we recommend an additional validation with per-class counting metrics (optional) and multi-threshold metrics (always recommended).
Select per-class counting metric (if any): If a decision rule applied to the predicted class scores is available (FP2.6), a per-class counting metric, such as the Fβ Score, should be selected. Each class of interest is separately assessed, preferably in a “one-versus-rest” fashion. The choice depends primarily on the decision rule and the distribution of classes (FP4.2). Details can be found in Subprocess S3 for selecting per-class counting metrics (Extended Data Fig. 3).
Select multi-threshold metric (if any): Counting metrics reduce the potentially complex output of a classifier (the continuous class scores) to a single value (the predicted class), such that they can work with a fixed confusion matrix. To compensate for this loss of information and obtain a more comprehensive picture of a classifier’s discriminatory performance, multi-threshold metrics work with a dynamic confusion matrix reflecting a range of possible thresholds applied to the predicted class scores. While we recommend the popular, well-interpretable and prevalence-independent AUROC as the default multi-threshold metric for classification, Average Precision (AP) can be more suitable in the case of high class balance because it incorporates predicted values, as detailed in Subprocess S4 for selecting multi-threshold metrics (Extended Data Fig. 4).
Select calibration metric (if any): If calibration assessment is requested (FP2.7), one or multiple calibration metrics should be added to the metric pool as detailed in Subprocess S5 for selecting calibration metrics (Extended Data Fig. 5).
Semantic segmentation (SemS).
In semantic segmentation, classification occurs at pixel level. However, it is not advisable to simply apply the standard classification metrics to the entire collection of pixels in a data set for two reasons. Firstly, pixels of the same image are highly correlated. Hence, to respect the hierarchical data structure, metric values should first be computed per image and then be aggregated over the set of images. Note in this context that the commonly used DSC is mathematically identical to the popular F1 Score applied at pixel level. Secondly, in segmentation problems, the user typically has an inherent interest in structure boundaries, centers or volumes of structures (FP2.1, FP2.2, FP2.3). The family of boundary-based metrics (subset of distance-based metrics) therefore requires the extraction of structure boundaries from the binary segmentation masks as a foundation for segmentation assessment. Based on these considerations and given all the complementary strengths and weaknesses of common segmentation metrics [65], we recommend the following process for segmentation problems (yellow path in Fig. 2; detailed description in Suppl. Note 2.3):
Select overlap-based metric (if any): In segmentation problems, counting metrics such as the DSC or IoU measure the overlap between the reference annotation and the algorithm prediction. As they can be considered the de facto standard for assessing segmentation quality and are well-interpretable, we recommend using them by default unless the target structures are consistently small, relative to the grid size (FP3.1), and the reference may be noisy (FP4.3.1). Depending on the specific properties of the problems, we recommend the DSC or IoU (default recommendation), the Fβ Score (preferred when there is a preference for either False Positive (FP) or False Negative (FN)) or the centerline Dice Similarity Coefficient (clDice) (for tubular structures). Details can be found in Subprocess S6 for selecting overlap-based metrics (Extended Data Fig. 6).
Select boundary-based metric (if any): Key weaknesses of overlap-based metrics include shape unawareness and limitations when dealing with small structures or high size variability [65]. Our general recommendation is therefore to complement an overlap-based metric with a boundary-based metric. If annotation imprecisions should be compensated for (FP2.5.7), our default recommendation is the Normalized Surface Distance (NSD). Otherwise, the fundamental user preference guiding metric selection is whether errors should be penalized by existence or distance (FP2.5.6), as detailed in Subprocess S7 for selecting boundary-based metrics (Extended Data Fig. 7).
Object detection (ObD).
Object detection problems differ from segmentation problems in several key features with respect to metric selection. Firstly, they involve distinguishing different instances of the same class and thus require the step of locating objects and assigning them to the corresponding reference object. Secondly, the granularity of localization is comparatively rough, which is why no boundary-based metrics are required (otherwise the problem would be phrased as an instance segmentation problem). Finally, and crucially important from a mathematical perspective, the absence of True Negatives (TNs) in object detection problems renders many popular classification metrics (e.g. Accuracy, Specificity, AUROC) invalid. In binary problems, for example, suitable counting metrics can only be based on three of the four entries of the confusion matrix. Based on these considerations and taking into account all the complementary strengths and weaknesses of existing metrics [65], we propose the following steps for object detection problems (green path in Fig. 2; detailed description in Suppl. Note 2.4):
Select localization criterion: An essential part of the validation is to decide whether a prediction matches a reference object. To this end, (1) the location of both the reference objects and the predicted objects must be adequately represented (e.g., by masks, bounding boxes or center points), and (2) a metric for deciding on a match (e.g. Mask IoU) must be chosen. As detailed in Subprocess S8 for selecting the localization criterion (Extended Data Fig. 8), our recommendation considers both the granularity of the provided reference (FP4.4) and the required granularity of the localization (FP2.4).
Select assignment strategy: As the localization does not necessarily lead to unambiguous matchings, an assignment strategy needs to be chosen to potentially resolve ambiguities that occurred during localization. As detailed in Subprocess S9 for selecting the assignment strategy (Extended Data Fig. 9), the recommended strategy depends on the availability of continuous class scores (FP5.1) as well as on whether double assignments should be punished (FP2.5.8).
Select classification metric(s) (if any): Once objects have been located and assigned to reference objects, generation of a confusion matrix (without TN) is possible. The final step therefore simply comprises choosing suitable classification metrics for validation. Several subfields of biomedical image analysis have converged to choosing solely a counting metric, such as the Fβ Score, as primary metric in object detection problems. We follow this recommendation when no continuous class scores are available for the detected objects (FP5.1). Otherwise, we disagree with the practice of basing performance assessment solely on a single, potentially suboptimal cutoff on the continuous class scores. Instead, we follow the recommendations for image-level classification and propose complementing a counting metric (Subprocess S3, Extended Data Fig. 3) with a multi-threshold metric (Subprocess S4, Extended Data Fig. 4) to obtain a more holistic picture of performance. As multi-threshold metric, we recommend AP or Free-Response Receiver Operating Characteristic (FROC) Score, depending on whether an easy interpretation (FROC Score) or a standardized metric (AP) is preferred. The choice of per-class counting metric depends primarily on the decision rule (FP2.6).
Note that the previous description implicitly assumed single-class problems, but generalization to multi-class problems is straightforward by applying the validation per-class. It is further worth mentioning that metric application is not straightforward in object detection problems as the number of objects in an image may be extremely small, or even zero, compared to the number of pixels in an image. Special considerations with respect to aggregation must therefore be made, as detailed in Suppl. Note 2.4.
Instance segmentation (InS).
Instance segmentation delivers the tasks of object detection and semantic segmentation at the same time. Thus, the pitfalls and recommendations for instance segmentation problems are closely related to those for segmentation and object detection [65]. This is directly reflected in our metric selection process (purple path in Fig. 2; detailed description in Suppl. Note 2.5):
Select object detection metric(s): To overcome problems related to instance unawareness (Fig. 1a, top left), we recommend selection of a set of detection metrics to explicitly measure detection performance. To this end, we recommend almost the exact process as for object detection with two exceptions. Firstly, given the fine granularity of both the output and the reference annotation, our recommendation for the localization strategy differs, as detailed in Subprocess S8 (Extended Data Fig. 8). Secondly, as depicted in S3 (Extended Data Fig. 3), we recommend the Panoptic Quality (PQ) [34] as an alternative to the Fβ Score. This metric is especially suited for instance segmentation, as it combines the assessment of overall detection performance and segmentation quality of successfully matched (True Positive (TP)) instances in a single score.
Select segmentation metric(s) (if any): In a second step, metrics to explicitly assess the segmentation quality for the TP instances may be selected. Here, we follow the exact same process as in semantic segmentation (Subprocesses S6, Extended Data Fig. 6 and S7, Extended Data Fig. 7). The primary difference is that the segmentation metrics are applied per-instance.
Importantly, the development process of the Metrics Reloaded framework was designed such that the pitfalls identified in the sister publication of this work [66] are comprehensively addressed. Tab. 1 makes the recommendations and design decisions corresponding to specific pitfalls explicit.
Once common reference-based metrics have been selected and, where necessary, complemented by application-specific metrics, the user proceeds with the application of the metrics to the given problem.
Step 3 - Metric Application.
Although the application of a metric to a given data set may appear straightforward, numerous pitfalls can occur [65]. Our recommendations for addressing them are provided in Extended Data Tab. 1. Following the taxonomy provided in the sister publication of this work [66], they are categorized in recommendations related to metric implementation, aggregation, ranking, interpretation, and reporting. While several aspects are covered in related work (e.g. [88]), an important contribution of the present work is the metric-specific summary of recommendations captured in the Metric Cheat Sheets (Suppl. Note 3.1). A further major contribution is our implementation of all Metrics Reloaded metrics in the open-source framework Medical Open Network for Artificial Intelligence (MONAI), available at https://github.com/Project-MONAI/MetricsReloaded (see Suppl. Methods).
Table 1. Metrics Reloaded addresses common and rare pitfalls in metric selection, as compiled in [66].
The first column lists all pitfall sources captured by the published taxonomy that relate to either the inadequate choice of the problem category or poor metric selection. The second column summarizes how Metrics Reloaded addresses these pitfalls. The notation FPX.Y refers to a fingerprint item (Suppl. Note 1.3).
| Source of Pitfall | Addressed in Metrics Reloaded by |
|---|---|
| Inadequate choice of the problem category | |
| Wrong choice of problem category | Problem category mapping (Subprocess S1, Fig. 4) as a prerequisite for metric selection. |
| Disregard of the domain interest | |
| Importance of structure boundaries | FP2.1 - Particular importance of structure boundaries; recommendation to complement common overlap-based segmentation metrics with boundary-based metrics (Fig. 2, Suppl. Note 2.3) if the property holds. |
| Importance of structure volume | FP2.2 - Particular importance of structure volume; recommendation to complement common overlap-based and boundary-based segmentation metrics with volume-based metrics (see Suppl. Note 2.3) if the property holds. |
| Importance of structure center(line) | FP2.3 - Particular importance of structure center(line); recommendation of the centerline Dice Similarity Coefficient (clDice) as alternative to the common Dice Similarity Coefficient (DSC) or Intersection over Union (IoU) in segmentation problems (Subprocess S6, Extended Data Fig. 6) and recommendation of center point-based localization criterion in object detection (Subprocess S8, Extended Data Fig. 8) if the property holds. |
| Importance of confidence awareness | FP2.7.1 - Calibration assessment requested; dedicated recommendations on calibration (Suppl. Note 2.6). |
| Importance of comparability across data sets | FP4.2 - Provided class prevalences reflect the population of interest; used in the Subprocesses S2–S4 (Extended Data Figs. 2–4); general focus on prevalence dependency of metrics in the framework. |
| Unequal severity of class confusions | FP2.5 - Penalization of errors; recommendation of the so far uncommon metric Expected Cost (EC) as classification metric (Subprocess S2, Extended Data Fig. 2); setting β in the Fβ Score according to preference for False Positive (FP) (oversegmentation) and False Negative (FN) (undersegmentation) (see DG3.3 in Suppl. Note 2.7.2). |
| Importance of cost-benefit-analysis | FP2.6 - Decision rule applied to predicted class scores: incorporation of a decision rule that is based on cost-benefit analysis; recommendation of the so far uncommon metrics Net Benefit (NB) (Fig. SN 3.11) and EC (Fig. SN 3.6). |
| Disregard of target structure properties | |
| Small structure sizes | FP3.1 - Small size of structures relative to pixel size; recommendation to consider the problem an object detection problem (Suppl. Note 2.4); complementation of overlap-based segmentation metrics with boundary-based metrics in the case of small structures with noisy reference (Subprocess S6, Extended Data Fig. 6); recommendation of lower object detection localization threshold in case of small sizes (see DG8.3 in Suppl. Note 2.7.7). |
| High variability of structure sizes | FP3.2 - High variability of structure sizes; recommendation of lower object detection localization threshold (see DG8.3 in Suppl. Note 2.7.7) and size stratification (Suppl. Note 2.4) in case of size variability. |
| Complex structure shapes | FP3.3 - Target structures feature tubular shape; recommendation of the clDice as alternative to the common DSC in segmentation problems (Subprocess S6, Extended Data Fig. 6) and recommendation of Point inside Mask/Box/Approx as localization criterion in object detection if the property holds (Subprocess S8, Extended Data Fig. 8). |
| Occurrence of overlapping or touching structures | FP3.5 - Possibility of overlapping or touching target structures; explicit recommendation to phrase problem as instance segmentation rather than semantic segmentation problem (Suppl. Note 2.3); recommendation of higher object detection localization threshold in case of small sizes (see DG8.3 in Suppl. Note 2.7.7). |
| Occurrence of disconnected structures | FP3.6 - Possibility of disconnected target structure(s); recommendation of appropriate localization criterion for object detection (DG8.2 in Suppl. Note 2.7.7). |
| Disregard of data set properties | |
| High class imbalance | FP4.1 - High class imbalance and FP2.5.5 - Compensation for class imbalances requested; compensation of class imbalance via prevalence-independent metrics such as EC and Balanced Accuracy (BA). |
| Small test set size | Recommendation of confidence intervals for all metrics. |
| Imperfect reference standard: Noisy reference standard | FP4.3.1 - High inter-rater variability and FP2.5.7 - Compensation for annotation imprecisions requested; default recommendation of the so far rather uncommon metric Normalized Surface Dice (NSD) to assess the quality of boundaries. |
| Imperfect reference standard: Spatial outliers in reference | FP4.3.2 - Possibility of spatial outliers in reference annotation and FP2.5.6 - Handling of spatial outliers; recommendation of outlier-robust metrics, such as NSD in case no distance-based penalization of outliers is requested in segmentation problems. |
| Occurrence of cases with an empty reference | FP4.6 - Possibility of reference without target structure(s); recommendations for aggregation in the case of empty references according to Suppl. Note 2.4 and Extended Data Tab.1. |
| Disregard of algorithm output properties | |
| Possibility of empty prediction | FP5.2 - Possibility of algorithm output not containing the target structure(s); selection of appropriate aggregation strategy in object detection (Suppl. Note 2.4). |
| Possibility of overlapping predictions | FP5.4 - Possibility of overlapping predictions; recommendation of an assignment strategy based on IoU > 0.5 if overlapping predictions are not possible and no predicted class scores are available. |
| Lack of predicted class scores | FP5.1 - Availability of predicted class scores; leveraging class scores for optimizing decision regions (FP2.6) and assessing calibration quality (FP2.7). |
Metrics Reloaded is broadly applicable in biomedical image analysis
To validate the Metrics Reloaded framework, we used it to generate recommendations for common use cases in biomedical image processing (see Suppl. Note 4). The traversal through the decision tree of our framework is detailed for eight selected use cases corresponding to the four different problem categories (Fig. 5):
Image-level classification (Figs. SN 5.5 - SN 5.8): frame-based sperm motility classification from time-lapse microscopy video of human spermatozoa (ImLC-1) and disease classification in dermoscopic images (ImLC-2).
Semantic segmentation (Figs. SN 5.9 - SN 5.10): embryo segmentation in microscopy images (SemS-1) and liver segmentation in Computed Tomography (computed tomography (CT)) images (SemS-2).
Object detection (Figs. SN 5.6 - SN 5.7, SN 5.11 - SN 5.12): cell detection and tracking during the autophagy process in time-lapse microscopy (ObD-1) and multiple sclerosis (MS) lesion detection in multi-modal brain magnetic resonance imaging (MRI) images (ObD-2).
Instance segmentation (Figs. SN 5.6 - SN 5.7, SN 5.9 - SN 5.12): instance segmentation of neurons from the fruit fly in 3D multi-color light microscopy images (InS-1) and surgical instrument instance segmentation in colonoscopy videos (InS-2).
The resulting metric recommendations (Fig. 5) demonstrate that a common framework across domains is sensible. In the showcased examples, shared properties of problems from different domains result in almost identical recommendations. In the semantic segmentation use cases, for example, the specific image modality is irrelevant for metric selection. What matters is the fact that a single object with a large size relative to the grid size should be segmented – properties that are captured by the proposed fingerprint. In Suppl. Note 4, we present recommendations for several other biomedical use cases.
The Metrics Reloaded online tool allows user-friendly metric selection
Selecting appropriate validation metrics while considering all potential pitfalls that may occur is a highly complex process, as demonstrated by the large number of figures in this paper. Some of the complexity, however, also results from the fact that the figures need to capture all possibilities at once. For example, many of the figures could be simplified substantially for problems based on only two classes. To leverage this potential and to improve the general user experience with our framework, we developed the Metrics Reloaded online tool, which is currently available as a beta version with restricted access (see Suppl. Methods). The tool captures our framework in a user-centric manner and can serve as a trustworthy common access point for image analysis validation.
DISCUSSION
Conventional scientific practice often grows through historical accretion, leading to standards that are not always well-justified. This holds particularly true for the validation standards in biomedical image analysis.
The present work represents the first comprehensive investigation and, importantly, constructive set of recommendations challenging the state of the art in biomedical image analysis algorithm validation with a specific focus on metrics. With the intention of revisiting – literally “re-searching” – common validation practices and developing better standards, we brought together experts from traditionally disjunct fields to leverage distributed knowledge. Our international consortium of more than 70 experts from the fields of biomedical image analysis, machine learning, statistics, epidemiology, biology, and medicine, representing a large number of relevant biomedical imaging initiatives and societies, developed the Metrics Reloaded framework that offers guidelines and tools to choose performance metrics in a problem-aware manner. The expert consortium was primarily compiled in a way to cover the required expertise from various fields but also consisted of researchers of different countries, (academic) ages, roles, and backgrounds (details can be found in the Suppl. Methods). Importantly, Metrics Reloaded comprehensively addresses all pitfalls related to metric selection (Tab. 1) and application (Extended Data Tab. 1) that were identified in this work’s sister publication [66].
Metrics Reloaded is the result of a 2.5-year long process involving numerous workshops, surveys, and expert group meetings. Many controversial debates were conducted during this time. Even deciding on the exact scope of the paper was anything but trivial. Our consortium eventually agreed on focusing on biomedical classification problems with categorical reference data and thus exploiting synergies across classification scales. Generating and handling fuzzy reference data (e.g., from multiple observers) is a topic of its own [45, 78] and was decided to be out of scope for this work. Furthermore, the inclusion of calibration metrics in addition to discrimination metrics was originally not intended because calibration is a complex topic in itself, and the corresponding field is relatively young and currently highly dynamic. This decision was reversed due to high demand from the community, expressed through crowdsourced feedback on the framework.
Extensive discussions also evolved around the inclusion criteria for metrics, considering the tradeoff between established (potentially flawed) and new (not yet stress-tested) metrics. Our strategy for arriving at the Metrics Reloaded recommendations balanced this tradeoff by using common metrics as a starting point and making adaptations where needed. For example, Weighted Cohen’s Kappa (WCK), originally designed for assessing inter-rater agreement, is the state-of-the-art metric used in the medical imaging community when handling ordinal data. Unlike other common multi-class metrics, such as (Balanced) Accuracy or MCC, it allows the user to specify different costs for different class confusions, thereby addressing the ordinal rating. However, our consortium deemed the (not widely known) metric EC generally more appropriate due to its favorable mathematical properties. Importantly, our framework does not intend to impose recommendations or act as a “black box”; instead, it enables users to make educated decisions while considering ambiguities and tradeoffs that may occur. This is reflected by our use of decision guides (Suppl. Note 2.7), which actively involve users in the decision-making process (for the example above, for instance, see DG2.1).
An important further challenge that our consortium faced was how to best provide recommendations in case multiple questions are asked for a single given data set. For example, a clinician’s ultimate interest may lie in assessing whether tumor progress has occurred in a patient. While this would be phrased as an image-level classification task (given two images as input), an interesting surrogate task could be seen in a segmentation task assessing the quality of tumor delineation and providing explainability for the results. Metrics Reloaded addresses the general challenge of multiple different driving biomedical questions corresponding to one data set pragmatically by generating a recommendation separately for each question. The same holds true for multi-label problems, for example, when multiple different types of abnormalities potentially co-occur in the same image/patient.
Another key challenge we faced was the validation of our framework due to the lack of ground truth “best metrics” to be applied for a given use case. Our solution builds upon three pillars. Firstly, we adopted established consensus building approaches utilized for developing widely used guidelines such as CONSORT [71], TRIPOD [55], or STARD [6]). Secondly, we challenged our initial recommendation framework by acquiring feedback via a social media campaign. Finally, we instantiated the final framework to a range of different biological and medical use cases. Our approach showcases the benefit of crowdsourcing as a means of expanding the horizon beyond the knowledge peculiar to specific scientific communities. The most prominent change effected in response to the social media feedback was the inclusion of the aforementioned EC, a powerful metric from the speech recognition community. Furthermore, upon popular demand, we added recommendations on assessing the interpretability of model outputs, now captured by Subprocess S5 (Extended Data Fig. 5).
After many highly controversial debates, the consortium ultimately converged on a consensus recommendation, as indicated by the high agreement in the final Delphi process (median agreement with the Subprocesses: 93%). While some subsprocesses (S1, S7, S8) were unanimously agreed on without a single negative vote, several issues were raised by individual researchers. While most of them were minor (e.g., concerning wording), a major debate revolved around calibration metrics. Some members, for example, questioned the value of stand-alone calibration metrics altogether. The reason for this view is the critically important misconception that the predicted class scores of a well-calibrated model express the true posterior probability of an input belonging to a certain class [62] – e.g., a patient’s risk for a certain condition based on an image. As this is not the case, several researchers argued for basing calibration assessment solely on proper scoring rules (such as the Brier Score (BS)), which assess the quality of the posteriors better than the stand-alone calibration metrics. We have addressed all these considerations in our recommendation framework including a detailed rationale for our recommendations, provided in Suppl. Note 2.6.
While we believe our framework to cover the vast majority of biomedical image analysis use cases, suggesting a comprehensive set of metrics for every possible biomedical problem may be out of its scope. The focus of our framework lies in correcting poor practices related to the selection of common metrics. However, in some use cases, common reference-based metrics – as a matter of principle – be unsuitable. In fact, the use of application-specific metrics may be required in some cases. A prominent example are instance segmentation problems in which the matching of reference and predicted instances is infeasible, causing overlap-based localization criteria to fail. Metrics such as Rand Index (RI) [63] and Variation of Information (VoI) [53] address this issue by avoiding one-to-one correspondences between predicted and reference instances. To make our framework applicable to such specific use cases, we integrated the step of choosing application-specific metrics in the main workflow (Fig. 2). Examples of such application-specific metrics can be found in related work [17, 20].
Metrics Reloaded primarily provides guidance for the selection of metrics that measure some notion of the “correctness” of an algorithm’s predictions on a set of test cases. It should be noted that holistic algorithm performance assessment also includes other aspects. One of them is robustness. For example, the accuracy of an algorithm for detecting disease in medical scans should ideally be the same across different hospitals that may use different acquisition protocols or scanners from different manufacturers. Recent work, however, shows that even the exact same models with nearly identical test set performance in terms of predictive accuracy may behave very differently on data from different distributions [18].
Reliability is another important algorithmic property to be taken into account during validation. A reliable algorithm should have the ability to communicate its confidence and raise a flag when the uncertainty is high and the prediction should be discarded [70]. For calibrated models, this can be achieved via the predicted class scores, although other methods based on dedicated model outputs trained to express the confidence or on density estimation techniques are similarly popular. Importantly, an algorithm with reliable uncertainty estimates or increased robustness to distribution shift might not always be the best performing in terms of predictive performance [28]. For safe use of classification systems in practice, careful balancing of the tradeoff between robustness and reliability over accuracy might be necessary.
So far, Metrics Reloaded focuses on common reference-based methods that compare model outputs to corresponding reference annotations. We made this design choice due to our hypothesis that reference-based metrics can be chosen in a modality- and application-agnostic manner using the concept of problem fingerprinting. As indicated by the step of choosing potential non-reference-based metrics (Fig. 2), however, it should be noted that validation and evaluation of algorithms should go far beyond purely technical performance [19, 80]. In this context, Jannin introduced a global concept of “responsible research” to encompass all possible high-level assessment aspects of a digital technology [30], including environmental, ethical, economical, social and societal aspects. For example, there are increasing efforts specifically devoted to the estimation of energy consumption and greenhouse gas emission of machine learning algorithms [39, 61, 76]. For these considerations, we would like to point the reader to available tools such as the Green Algorithms calculator [40] or Carbontracker [89].
It must further be noted that while Metrics Reloaded places a focus on the selection of metrics, adequate application is also important. Detailed failure case analysis [68] and performance assessment on relevant subgroups, for example, have been highlighted as critical components for better understanding when and where an algorithm may fail [10, 58]. Given that learning-based algorithms rely on the availability of historical data sets for training, there is a real risk that any existing biases in the data may be picked up and replicated or even exacerbated when an algorithm makes predictions [1, 22]. This is of particular concern in the context of systemic biases in healthcare, such as the scarcity of representative data from underserved populations and often higher error rates in diagnostic labels in particular subgroups [27, 59]. Relevant meta information such as patient demographics, including biological sex and ethnicity, needs to be accessible for the test sets such that potentially disparate performance across subgroups can be detected [52]. Here, it is important to make use of adequate aggregations over the validation metrics as disparities in minority groups might otherwise be missed.
Finally, it must be noted that our framework addresses metric choice in the context of technical validation of biomedical algorithms. For translation of an algorithm into, for example, clinical routine, this validation may be followed by a (clinical) validation step assessing its performance compared to conventional, non-algorithm-based care according to patient-related outcome measures, such as overall survival [60].
A key remaining challenge for Metric Reloaded is its dissemination such that it will substantially contribute to raising the quality of biomedical imaging research. To encourage widespread adherence to new standards, entry barriers should be as low as possible. While the framework with its vast number of subprocesses may seem very complex at first, it is important to note that from a user perspective only a fraction of the framework is relevant for a given task, making the framework more tangible. This is notably illustrated by the Metric Reloaded online tool, which substantially simplifies the metric selection procedure. As is common in scientific guideline and recommendation development, we intend to regularly update our framework to reflect current developments in the field, such as the inclusion of new metrics or biomedical use cases. This is intended to include an expansion of the framework’s scope to further problem categories, such as regression and reconstruction. In order to accommodate future developments in a fast and efficient manner, we envision our consortium building consensus through accelerated Delphi rounds organized by the Metric Reloaded core team. Once consensus is obtained, changes will be implemented in both the framework and online tool and highlighted so that users can easily identify changes to the previous version, which will ensure full transparency and comparability of results. In this way, we envision the Metrics Reloaded framework and online tool as a dynamic resource reliably reflecting the current state of the art at any given time point in the future, for years to come.
Of note, while the provided recommendations originate from the biomedical image analysis community, many aspects generalize to imaging research as a whole. Particularly the recommendations derived for individual fingerprints (e.g., implications of class imbalance) hold across domains, although it is possible that for different domains the existing fingerprints would need to be complemented by further features that this community is not aware of.
In conclusion, the Metrics Reloaded framework provides biomedical image analysis researchers with the first systematic guidance on choosing validation metrics across different imaging tasks in a problem-aware manner. Through its reliance on methodology that can be generalized, we envision the Metrics Reloaded framework to spark a scientific debate and hopefully lead to similar efforts being undertaken in other areas of imaging research, thereby raising research quality on a much larger scale than originally anticipated. In this context, our framework and the process by which it was developed could serve as a blueprint for broader efforts aimed at providing reliable recommendations and enforcing adherence to good practices in imaging research.
Extended Data
Extended Data Tab. 1. Recommendations for metric application addressing the pitfalls collected in [19].
The first column comprises all sources of pitfalls captured by the published taxonomy that relate to the application of (already selected) metrics. The second column provides the Metrics Reloaded recommendation. The notation FPX.Y refers to a fingerprint item (Suppl. Note 1.3).
| Source of Pitfall | Recommendation |
|---|---|
| Metric implementation | |
| Non-standardized metric definition and undefined corner cases | Use reference implementations provided at https://github.com/Project-MONAI/MetricsReloaded |
| Discretization issues | Use unbiased estimates of properties of interest if possible (Suppl. Note 2.6). |
| Metric-specific issues including sensitivity to hyperparameters | Read metric-specific recommendations in the cheat sheets (Suppl. Note 3.1). |
| Aggregation | |
| Hierarchical label/class structure | Address the potential correlation between classes when aggregating [Kang & Sukthankar, 2006]. |
| Multi-class problem | Complement validation with multi-class metrics such as Expected Cost (EC) or Matthews Correlation Coefficient (MCC) with per-class validation (Fig. 2); perform weighted class aggregation if FP2.5.1 Unequal interest across classes holds. |
| Non-independence of test cases (FP4.5) | Respect the hierarchical data structure when aggregating metrics [Liang & Zeger, 1986]. |
| Risk of bias | Leverage metadata (e.g. on imaging device/protocol/center) to reveal potential algorithmic bias [Badgeley et al., 2019]. |
| Possibility of invalid prediction (FP5.3) | Follow category-specific aggregation strategy detailed in Suppl. Note 2. |
| Ranking | |
| Metric relationships | Avoid combining closely related metrics (see Fig. SN 2.1) when choosing metrics to be used in algorithm ranking. |
| Ranking uncertainties | Provide information beyond plain tables that make possible uncertainties in rankings explicit as detailed in [30]. |
| Reporting | |
| Non-determinism of algorithms | Consider multiple test set runs to address the variability of results resulting from non-determinism [Khan et al., 2019, Summers & Dinneen, 2021]. |
| Uninformative visualization | Include a visualization of the raw metric values [30] and report the full confusion matrix unless FP2.6 = no decision rule applied holds. |
| Interpretation | |
| Low resolution | Read metric-related recommendations to obtain awareness of the pitfall (Suppl. Note 3.1). |
| Lack of lower/upper bounds | Read metric-related recommendations to obtain awareness of the pitfall (Suppl. Note 3.1). |
| Insufficient domain relevance of metric score differences | Report on the quality of the reference (e.g. intra-rater and inter-rater variability) [Kottner et al., 2011]. Choose the number of decimal places such that they reflect both relevance and uncertainties of the reference. More than one decimal number is often not useful given the typically high inter-rater variability. |
[Kang & Sukthankar, 2006] Kang, F., Jin, R., & Sukthankar, R. (2006, June). Correlated label propagation with application to multi-label learning. In 2006 IEEE Computer Society Conference on Computer Vision and Pattern Recognition (CVPR’06) (Vol. 2, pp. 1719–1726). IEEE.
[Liang & Zeger, 1986] Liang, K. Y., & Zeger, S. L. (1986). Longitudinal data analysis using generalized linear models. Biometrika, 73(1), 13–22.
[Badgeley et al., 2019] Badgeley, M. A., Zech, J. R., Oakden-Rayner, L., Glicksberg, B. S., Liu, M., Gale, W., … & Dudley, J. T. (2019). Deep learning predicts hip fracture using confounding patient and healthcare variables. NPJ digital medicine, 2(1), 31.
[Khan et al., 2019] Khan, D. A., Li, L., Sha, N., Liu, Z., Jimenez, A., Raj, B., & Singh, R. (2019). Non-Determinism in Neural Networks for Adversarial Robustness. arXiv preprint arXiv:1905.10906.
[Summers & Dinneen, 2021] Summers, C., & Dinneen, M. J. (2021, July). Nondeterminism and instability in neural network optimization. In International Conference on Machine Learning (pp. 9913–9922). PMLR.
[Kottner et al., 2011] Kottner, J., Audigé, L., Brorson, S., Donner, A., Gajewski, B. J., Hróbjartsson, A., … & Streiner, D. L. (2011). Guidelines for reporting reliability and agreement studies (GRRAS) were proposed. International journal of nursing studies, 48(6), 661–671.
Extended Data Fig. 1:
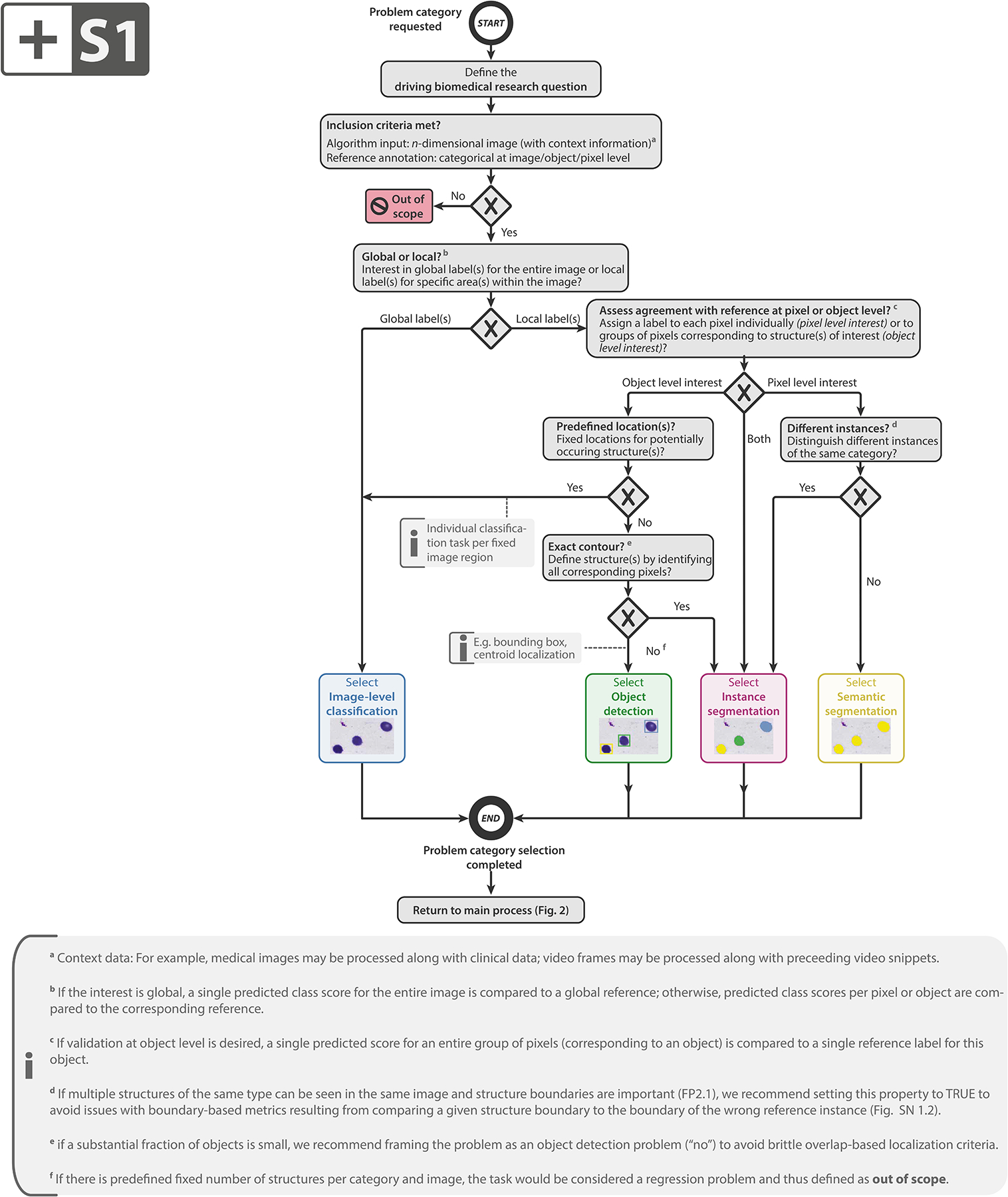
Subprocess S1 for selecting a problem category. Subprocess S1 for selecting a problem category. The Category Mapping maps a given research problem to the appropriate problem category with the goal of grouping problems by similarity of validation. The leaf nodes represent the categories: image-level classification, object detection, instance segmentation, or semantic segmentation. FP2.1 refers to fingerprint 2.1 (see Fig. SN 1.10). An overview of the symbols used in the process diagram is provided in Fig. SN 5.1.
Extended Data Fig. 2:
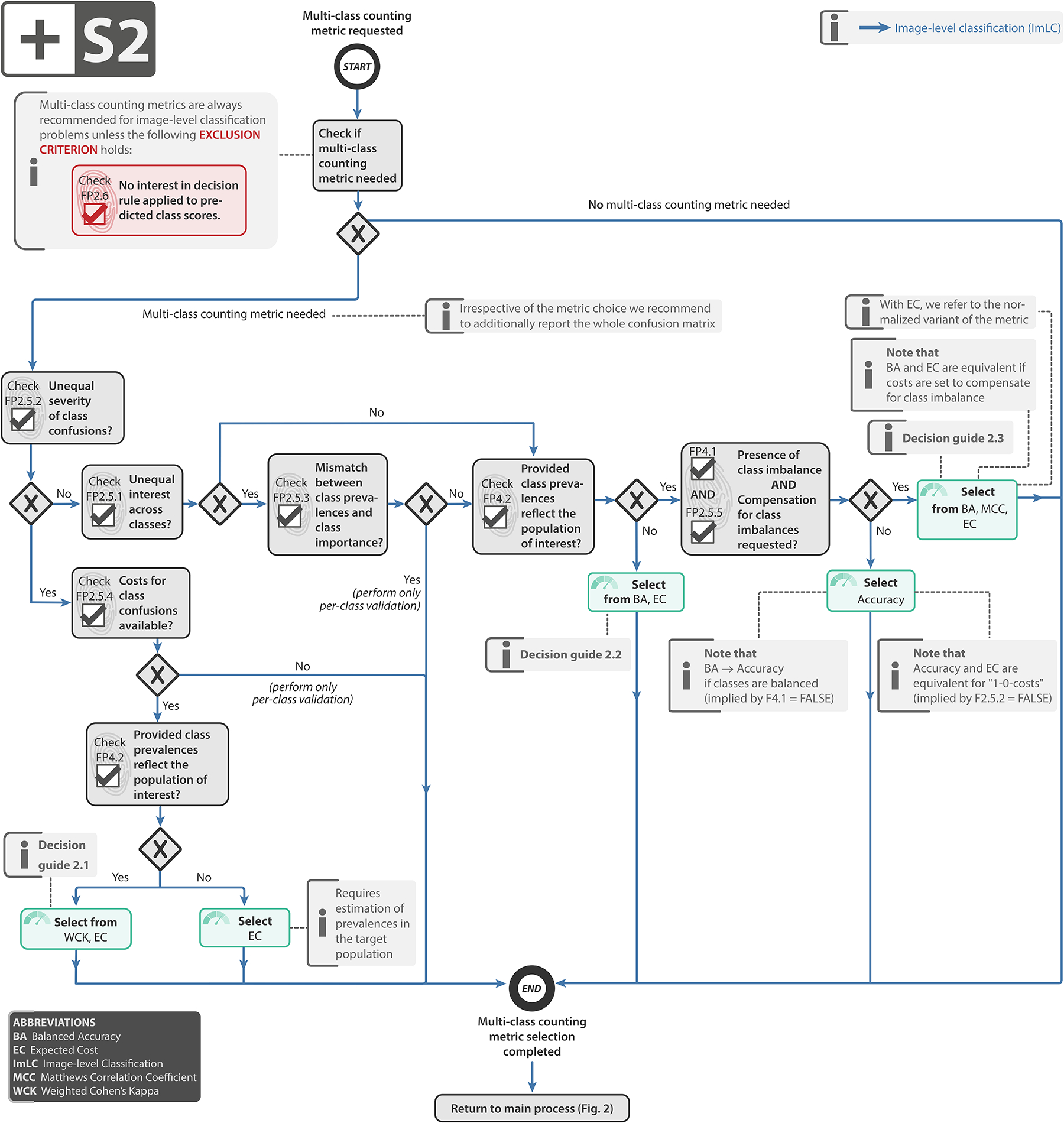
Subprocess S2 for selecting multi-class metrics (if any).Subprocess S2 for selecting multi-class metrics (if any). Applies to: image-level classification (ImLC). In the case of presence of class imbalance and no compensation of class imbalance being requested, one should follow the “No” branch. Decision guides are provided in Suppl. Note 2.7.1. A detailed description of the subprocess is given in Suppl. Note 2.2.
Extended Data Fig. 3:
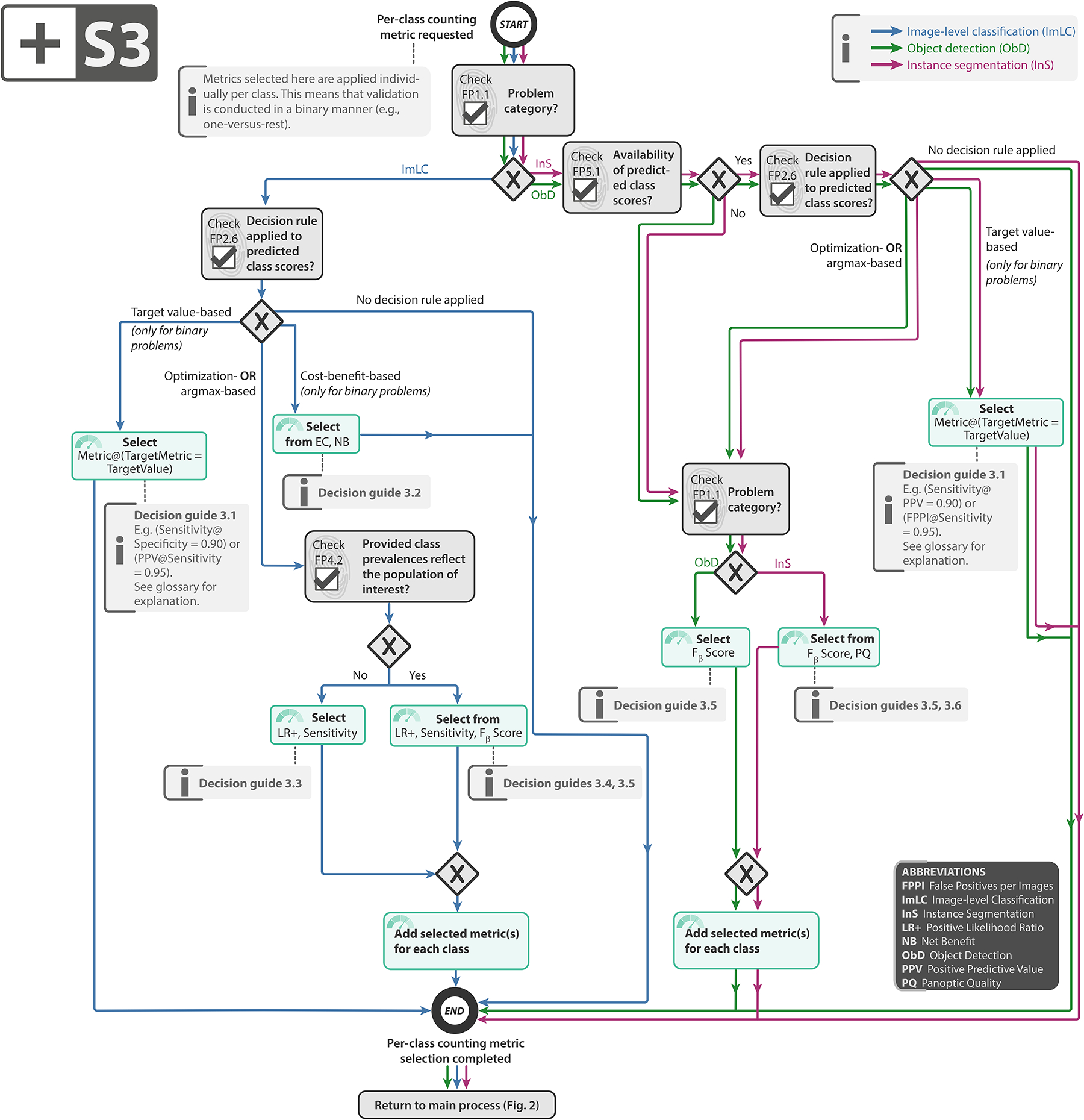
Subprocess S3 for selecting a per-class counting metric (if any). Subprocess S3 for selecting a per-class counting metric (if any). Applies to: image-level classification (ImLC), object detection (ObD), and instance segmentation (InS). Decision guides are provided in Suppl. Note 2.7.2. A detailed description of the subprocess is given in Suppl. Notes 2.2, 2.4, and 2.5.
Extended Data Fig. 4.
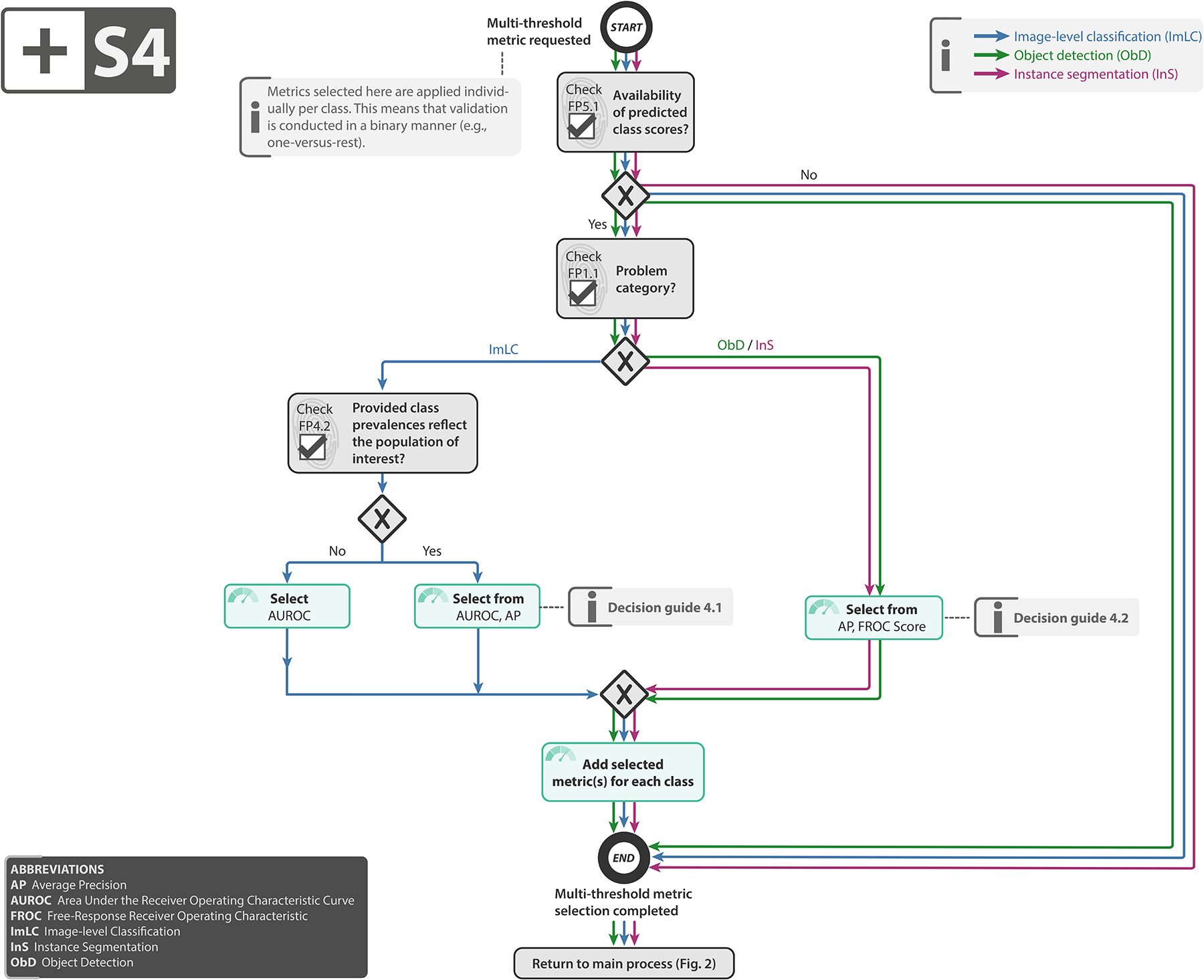
Subprocess S4 for selecting a multi-threshold metric (if any). Subprocess S4 for selecting a multi-threshold metric (if any). Applies to: image-level classification (ImLC), object detection (ObD), and instance segmentation (InS). Decision guides are provided in Suppl. Note 2.7.3. A detailed description of the subprocess is given in Suppl. Notes 2.2, 2.4, and 2.5.
Extended Data Fig. 5:
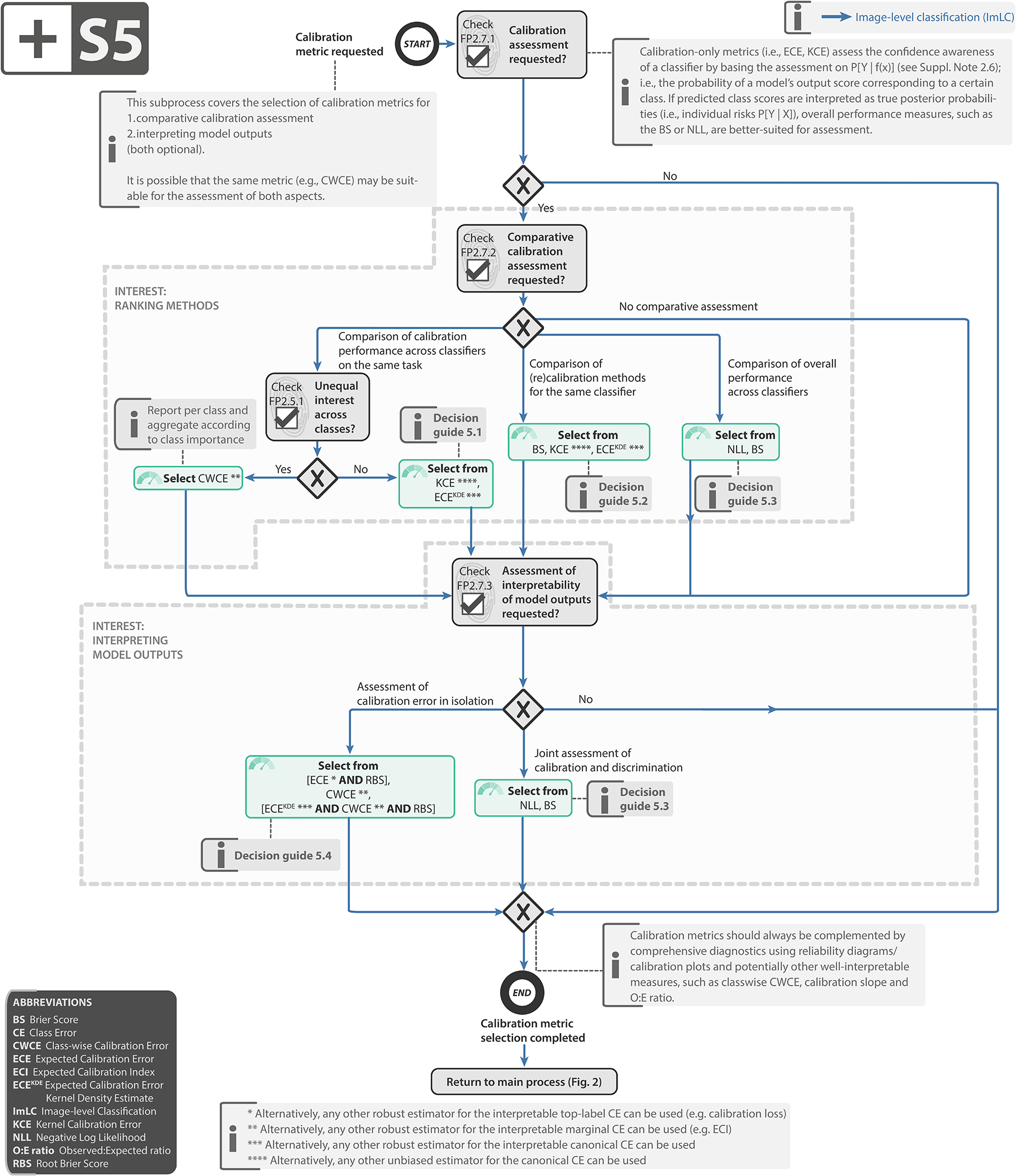
Subprocess S5 for selecting a calibration metric (if any). Subprocess S5 for selecting a calibration metric (if any). Applies to: image-level classification (ImLC). Decision guides are provided in Suppl. Note 2.7.4. A detailed description of the subprocess is given in Suppl. Note 2.6. Further suggested calibration metrics include the calibration loss [8], calibration slope [76], Expected Calibration Index (ECI) [87] and Observed:Expected ratio (O:E ratio) [68].
Extended Data Fig. 6:
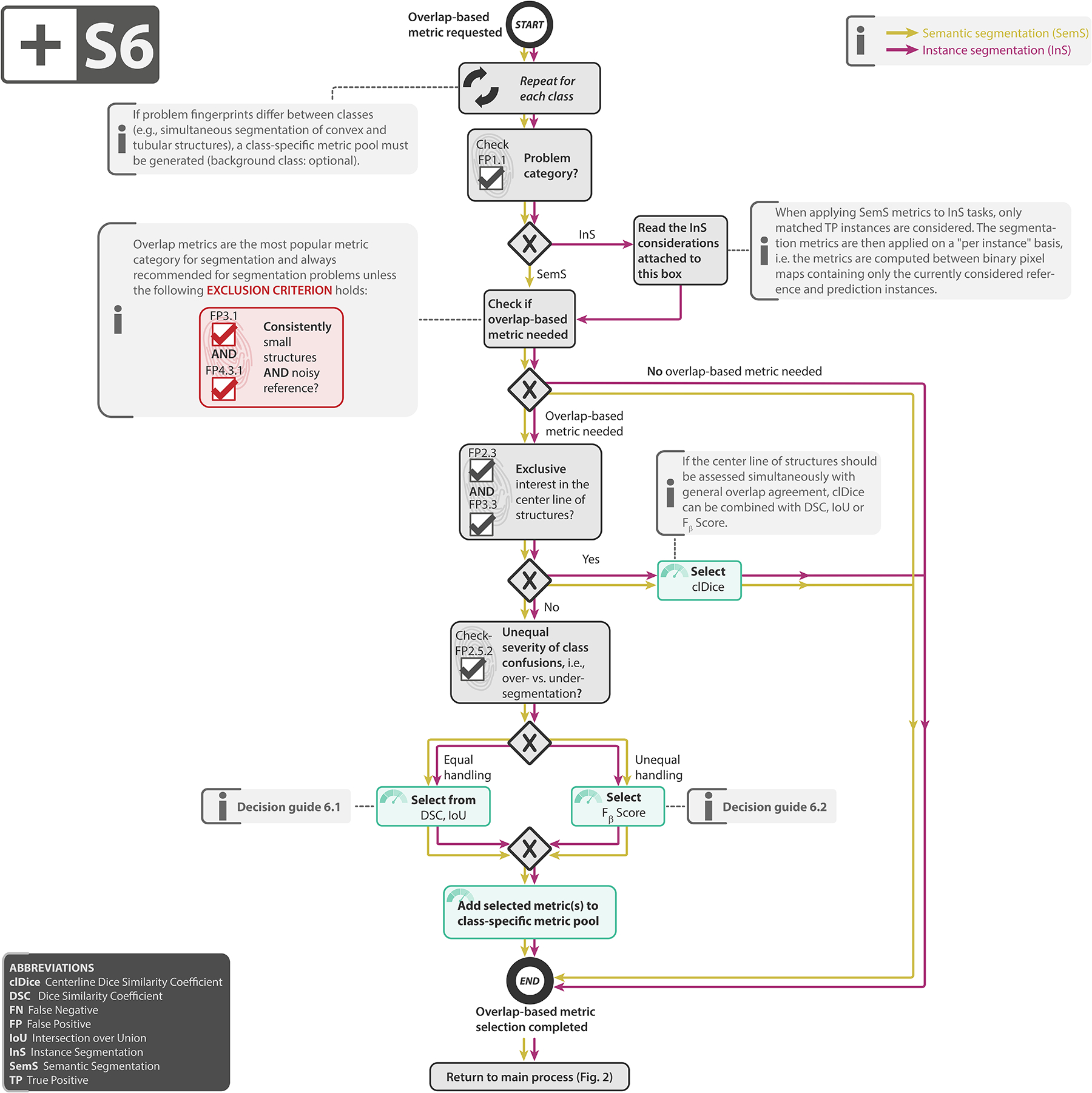
Subprocess S6 for selecting overlap-based segmentation metrics (if any). Subprocess S6 for selecting overlap-based segmentation metrics (if any). Applies to: semantic segmentation (SemS) and instance segmentation (InS). Decision guides are provided in Suppl. Note 2.7.5. A detailed description of the subprocess is given in Suppl. Notes 2.3 and 2.5.
Extended Data Fig. 7:
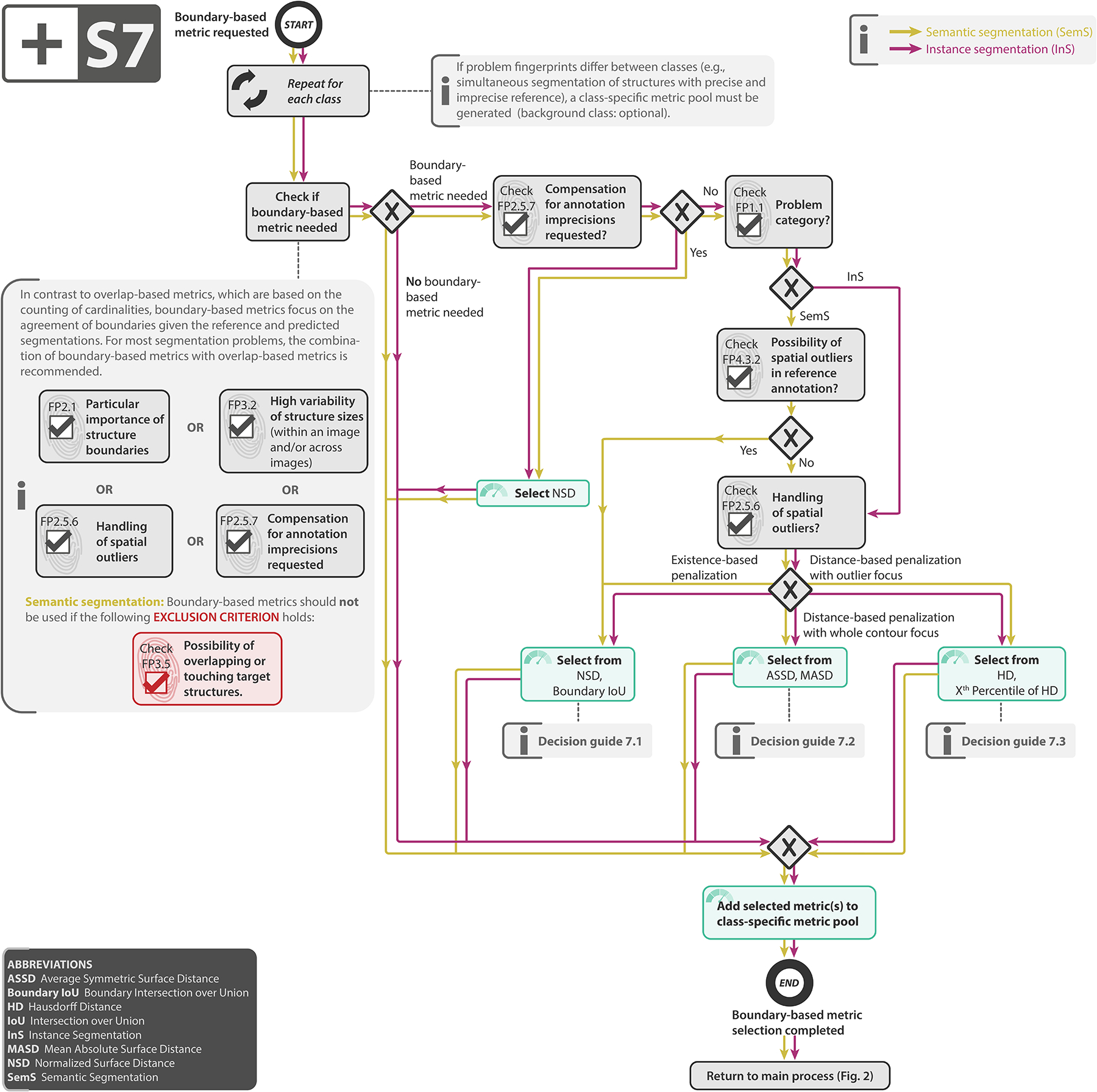
Subprocess S7 for selecting a boundary-based segmentation metric (if any). Subprocess S7 for selecting a boundary-based segmentation metric (if any). Applies to: semantic segmentation (SemS) and instance segmentation (InS). Decision guides are provided in Suppl. Note 2.7.6. A detailed description of the subprocess is given in Suppl. Notes 2.3 and 2.5.
Extended Data Fig. 8:
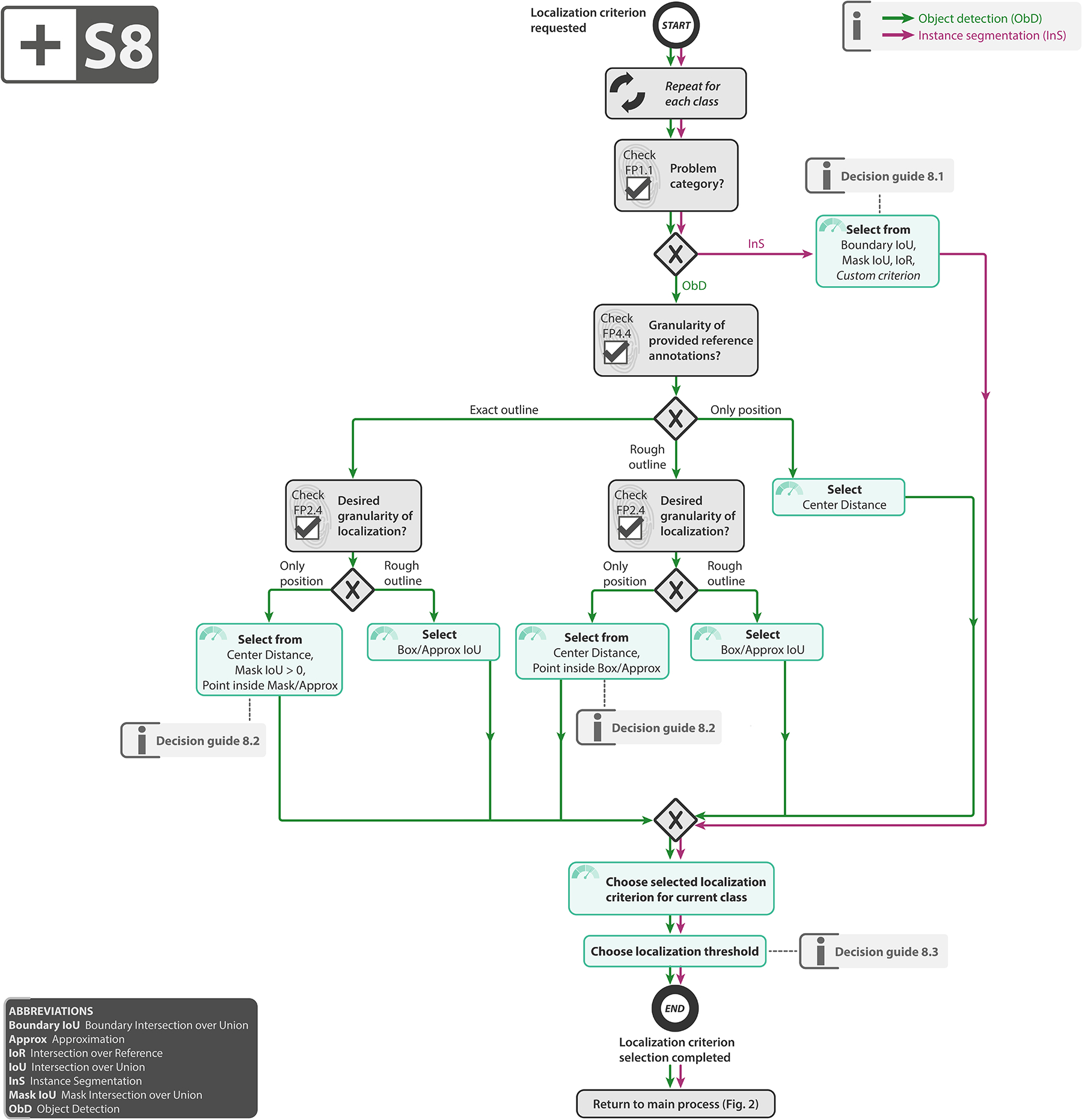
Subprocess S8 for selecting the localization criterion. Subprocess S8 for selecting the localization criterion. Applies to: object detection (ObD) and instance segmentation (InS). Definitions of the localization criteria can be found in [66]. Decision guides are provided in Suppl. Note 2.7.7. A detailed description of the subprocess is given in Suppl. Notes 2.4 and 2.5.
Extended Data Fig. 9:
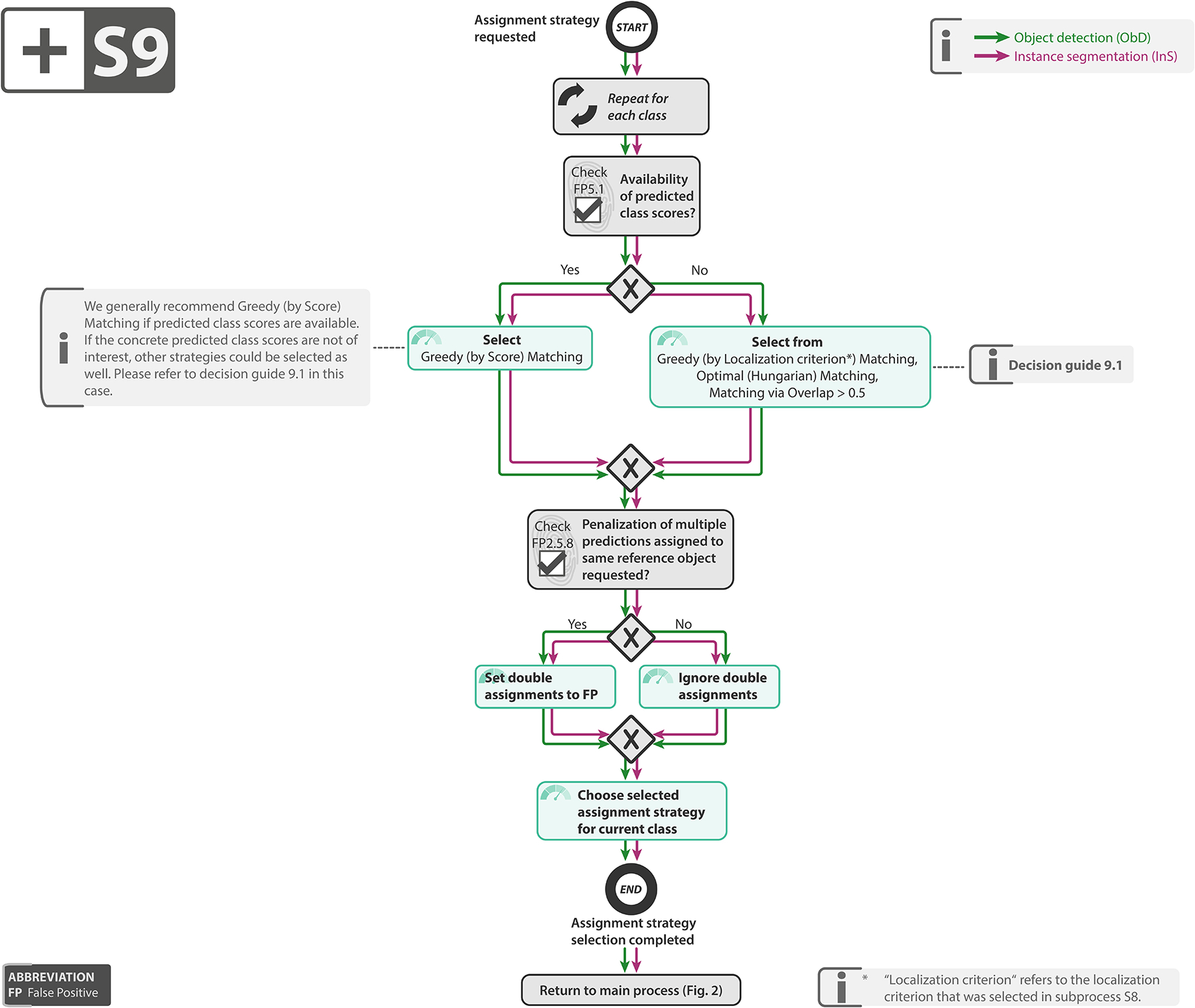
Subprocess S9 for selecting the assignment strategy. Subprocess S9 for selecting the assignment strategy. Applies to: object detection (ObD) and instance segmentation (InS). Assignment strategies are defined in [66]. Decision guides are provided in Suppl. Note 2.7.8. A detailed description of the subprocess is given in Suppl. Notes 2.4 and 2.5.
Supplementary Material
ACKNOWLEDGEMENTS
This work was initiated by the Helmholtz Association of German Research Centers in the scope of the Helmholtz Imaging Incubator (HI), the MICCAI Special Interest Group on biomedical image analysis challenges and the benchmarking working group of the MONAI initiative. It received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement No. [101002198], NEURAL SPICING). It was further supported in part by the Intramural Research Program of the National Institutes of Health (NIH) Clinical Center as well as by the National Cancer Institute (NCI) and the National Institute of Neurological Disorders and Stroke (NINDS) of the NIH, under award numbers NCI:U01CA242871 and NINDS:R01NS042645. The content of this publication is solely the responsibility of the authors and does not represent the official views of the NIH. T.A. acknowledges the Canada Institute for Advanced Research (CIFAR) AI Chairs program, the Natural Sciences and Engineering Research Council of Canada. F.B. was co-funded by the European Union (ERC, TAIPO, 101088594). Views and opinions expressed are however those of the authors only and do not necessarily reflect those of the European Union or the European Research Council. Neither the European Union nor the granting authority can be held responsible for them. V.C. acknowledges funding from NovoNordisk Foundation (NNF21OC0068816) and Independent Research Council Denmark (1134-00017B). B.A.C. was supported by NIH grant P41 GM135019 and grant 2020-225720 from the Chan Zuckerberg Initiative DAF, an advised fund of the Silicon Valley Community Foundation.
G.S.C. was supported by Cancer Research UK (programme grant: C49297/A27294). M.M.H. is supported by the Natural Sciences and Engineering Research Council of Canada (RGPIN-2022- 05134). A.Kara. is supported by French State Funds managed by the “Agence Nationale de la Recherche (ANR)” - “Investissements d’Avenir” (Investments for the Future), Grant ANR-10-IAHU-02 (IHU Strasbourg). M.K. was supported by the Ministry of Education, Youth and Sports of the Czech Republic (Project LM2018129). T.K. was supported in part by 4UH3-CA225021-03, 1U24CA180924- 01A1, 3U24CA215109-02, and 1UG3-CA225-021-01 grants from the National Institutes of Health.
G.L. receives research funding from the Dutch Research Council, the Dutch Cancer Association, HealthHolland, the European Research Council, the European Union, and the Innovative Medicine Initiative. C.H.S. is supported by an Alzheimer’s Society Junior Fellowship (AS-JF-17-011). M.R is supported by Innosuisse grant number 31274.1 and Swiss National Science Foundation Grant Number 205320_212939. R.M.S. is supported by the Intramural Research Program of the NIH Clinical Center. A.T. acknowledges support from Academy of Finland (Profi6 336449 funding program), University of Oulu strategic funding, Finnish Foundation for Cardiovascular Research, Wellbeing Services County of North Ostrobothnia (VTR project K62716), and Terttu foundation.
S.A.T. acknowledges the support of Canon Medical and the Royal Academy of Engineering and the Research Chairs and Senior Research Fellowships scheme (grant RCSRF1819\8\25).
We thank Nina Sautter, Patricia Vieten and Tim Adler for proposing the name for the project. We would like to thank Peter Bankhead, Fred Hamprecht, Hannes Kenngott, David Moher, and Bram Stieltjes for fruitful discussions on the framework.
We thank Susanne Steger for the data protection supervision and Anke Trotter for the hosting of the surveys.
We would like to thank Lisa Mais for instantiating the use case for instance segmentation of neurons from the fruit fly in 3D multicolor light microscopy images. We would further like to thank the Janelia FlyLight Project Team for providing us with example images for this use case.
We would like to thank the following people for testing the metric mappings, reviewing the recommendations and performing metric-centric testing: Tim Adler, Christoph Bender, Ahmad Bin Qasim, Kris Dreher, Niklas Holzwarth, Marco Hübner, Dominik Michael, Lucas-Raphael Müller, Maike Rees, Tom Rix, Melanie Schellenberg, Silvia Seidlitz, Jan Sellner, Akriti Srivastava, Fabian Wolf, Amine El Yamlahi, Silvia D. Almeida, Michael Baumgartner, Dimitrios Bounias, Till Bungert, Maximilian Fischer, Lukas Klein, Gregor Köhler, Bálint Kovács, Carsten Lueth, Tobias Norajitra, Constantin Ulrich, Tassilo Wald, Iuliia Alekseenko, Xiao Liu, Andrea Marheim Storås, Vajira Thambawita.
We would like to thank the following people for taking our social media community survey and providing helpful feedback for improving the framework: Yamashita Akemi, Roi Anteby, Callum Arthurs, Pieter De Backer, Henry Badgery, Matthew Baugh, Jose Bernal, Matthew Blaschko, Dimitrios Bounias, Felipe Campos Kitamura, Jacob Carse, Chen Chen, Ivo Flipse, Nicolas Gaggion, Camila González, Pedro M. Gordaliza, Tim Horeman, Leo Joskowicz, Abin Jose, Amith Kamath, Brendan Kelly, Yannick Kirchhoff, Levin Arne Kobelke, Lars Krämer, Mira Krendel, John LaMaster, Thomas de Lange, Joël L. Lavanchy, Jianning Li, Carsten Lüth, Lisa Mais, Andrea Marheim Storås, Vishwesh Nath, Cian Scannell, Constantin Pape, M.P. Schijven, Alberto Selvanetti, Bella Specktor Fadida, Roger Staff, Jeremy Tan, Eric Tkaczyk, Rodrigo Tripodi Calumby, Athanasios Vlontzos, Weitong Zhang, Can Zhao, Jiayi Zhu.
Footnotes
CODE AVAILABILITY STATEMENT
We provide reference implementations for all Metrics Reloaded metrics within the MONAI open-source framework. They are accessible at https://github.com/Project-MONAI/MetricsReloaded.
COMPETING INTERESTS
The authors declare the following competing interests: Under his terms of employment, M.B.B. is entitled to stock options in Mona.health, a KU Leuven spinoff. F.B. is an employee of Siemens AG (Munich, Germany). F.B. reports funding from Merck (Darmstadt, Germany). B.v.G. is a shareholder of Thirona (Nijmegen, NL). B.G. was an employee of HeartFlow Inc (California, USA) and Kheiron Medical Technologies Ltd (London, UK). M.M.H. received an Nvidia GPU Grant. B.K. is a consultant for ThinkSono Ldt (London, UK). G.L. is on the advisory board of Canon Healthcare IT (Minnetonka, USA) and is a shareholder of Aiosyn BV (Nijmegen, NL). N.R. is an employee of Nvidia GmbH (Munich, Germany). J.S.-R. reports funding from GSK (Heidelberg, Germany), Pfizer (New York, USA) and Sanofi (Paris, France) and fees from Travere Therapeutics (California, USA), Stadapharm (Bad Vilbel, Germany), Astex Therapeutics (Cambridge, UK), Pfizer (New York, USA), and Grunenthal (Aachen, Germany). R.M.S. receives patent royalties from iCAD (New Hampshire, USA), ScanMed (Nebraska, USA), Philips (Amsterdam, NL), Translation Holdings (Alabama, USA) and PingAn (Shenzhen, China); his lab received research support from PingAn through a Cooperative Research and Development Agreement. S.A.T. receives financial support from Canon Medical Research Europe (Edinburgh, Scotland). The remaining authors declare no competing interests
Not to be confused with distance metrics in the pure mathematical sense.
We thank the Intelligent Medical Systems (IMSY) lab members Nina Sautter, Patricia Vieten and Tim Adler for the suggestion of the name, inspired by the Matrix movies.
DATA AVAILABILITY STATEMENT
No data was used in this study.
REFERENCES
- [1].Adamson Adewole S and Smith Avery. Machine learning and health care disparities in dermatology, 2018. [DOI] [PubMed] [Google Scholar]
- [2].Antonelli Michela, Reinke Annika, Bakas Spyridon, Farahani Keyvan, Kopp-Schneider Annette, Landman Bennett A, Litjens Geert, Menze Bjoern, Ronneberger Olaf, Summers Ronald M, et al. The medical segmentation decathlon. Nature Communications, 13(1):1–13, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Armato Samuel G III, McLennan Geoffrey, Bidaut Luc, McNitt-Gray Michael F, Meyer Charles R, Reeves Anthony P, Zhao Binsheng, Aberle Denise R, Henschke Claudia I, Hoffman Eric A, et al. The lung image database consortium (lidc) and image database resource initiative (idri): a completed reference database of lung nodules on ct scans. Medical physics, 38(2):915–931, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Badgeley MA, Zech JR, Oakden-Rayner L, Glicksberg BS, Liu M, Gale W, McConnell MV, Percha B, Snyder TM, and Dudley JT. Deep learning predicts hip fracture using confounding patient and healthcare variables. npj digit med. 2019; 2: 31, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Birhane Abeba, Kalluri Pratyusha, Card Dallas, Agnew William, Dotan Ravit, and Bao Michelle. The values encoded in machine learning research. arXiv, June 2021. [Google Scholar]
- [6].Bossuyt Patrick M, Reitsma Johannes B, Bruns David E, Gatsonis Constantine A, Glasziou Paul P, Irwig Les M, Lijmer Jeroen G, Moher David, Rennie Drummond, De Vet Henrica CW, et al. Towards complete and accurate reporting of studies of diagnostic accuracy: the stard initiative. Annals of internal medicine, 138(1):40–44, 2003. [DOI] [PubMed] [Google Scholar]
- [7].Brown Bernice B. Delphi process: a methodology used for the elicitation of opinions of experts. Technical report, Rand Corp; Santa Monica CA, 1968. [Google Scholar]
- [8].Brümmer Niko and Du Preez Johan. Application-independent evaluation of speaker detection. Computer Speech & Language, 20(2–3):230–275, 2006. [Google Scholar]
- [9].Carass Aaron, Roy Snehashis, Gherman Adrian, Reinhold Jacob C, Jesson Andrew, Arbel Tal, Maier Oskar, Handels Heinz, Ghafoorian Mohsen, Platel Bram, et al. Evaluating white matter lesion segmentations with refined sørensen-dice analysis. Scientific reports, 10(1):1–19, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Char Danton S, Shah Nigam H, and Magnus David. Implementing machine learning in health care - addressing ethical challenges. N. Engl. J. Med, 378(11):981–983, March 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Chenouard Nicolas, Smal Ihor, De Chaumont Fabrice, Maška Martin, Sbalzarini Ivo F, Gong Yuanhao, Cardinale Janick, Carthel Craig, Coraluppi Stefano, Winter Mark, et al. Objective comparison of particle tracking methods. Nature methods, 11(3):281–289, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Codella Noel, Rotemberg Veronica, Tschandl Philipp, Celebi M Emre, Dusza Stephen, Gutman David, Helba Brian, Kalloo Aadi, Liopyris Konstantinos, Marchetti Michael, et al. Skin lesion analysis toward melanoma detection 2018: A challenge hosted by the international skin imaging collaboration (isic). arXiv preprint arXiv:1902.03368, 2019. [Google Scholar]
- [13].Collins Gary S, Dhiman Paula, Andaur Navarro Constanza L, Ma Jie, Hooft Lotty, Reitsma Johannes B, Logullo Patricia, Beam Andrew L, Peng Lily, Van Calster Ben, et al. Protocol for development of a reporting guideline (tripod-ai) and risk of bias tool (probast-ai) for diagnostic and prognostic prediction model studies based on artificial intelligence. BMJ open, 11(7):e048008, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Commowick Olivier, Istace Audrey, Kain Michael, Laurent Baptiste, Leray Florent, Simon Mathieu, Pop Sorina Camarasu, Girard Pascal, Ameli Roxana, Ferré Jean-Christophe, et al. Objective evaluation of multiple sclerosis lesion segmentation using a data management and processing infrastructure. Scientific reports, 8(1):1–17, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].CONSORT-AI and SPIRIT-AI Steering Group. Reporting guidelines for clinical trials evaluating artificial intelligence interventions are needed. Nat. Med, 25(10):1467–1468, October 2019. [DOI] [PubMed] [Google Scholar]
- [16].Correia Paulo and Pereira Fernando. Video object relevance metrics for overall segmentation quality evaluation. EURASIP Journal on Advances in Signal Processing, 2006:1–11, 2006. [Google Scholar]
- [17].Côté Marc-Alexandre, Girard Gabriel, Boré Arnaud, Garyfallidis Eleftherios, Houde Jean-Christophe, and Descoteaux Maxime. Tractometer: towards validation of tractography pipelines. Medical Image Analysis, 17(7):844–857, October 2013. ISSN 1361–8423. doi: 10.1016/j.media.2013.03.009. [DOI] [PubMed] [Google Scholar]
- [18].D’Amour A, Heller K, Moldovan D, Adlam B, and others. Underspecification presents challenges for credibility in modern machine learning. arXiv preprint arXiv, 2020. [Google Scholar]
- [19].Université de Montréal. The Declaration - Montreal Responsible AI, 2017. URL https://www.montrealdeclaration-responsibleai.com/the-declaration. [Google Scholar]
- [20].Ellis David G, Alvarez Carlos M, and Aizenberg Michele R. Qualitative criteria for feasible cranial implant designs. In Cranial Implant Design Challenge, pages 8–18. Springer, 2021. [Google Scholar]
- [21].Ferrer Luciana. Analysis and comparison of classification metrics. arXiv preprint arXiv:2209.05355, 2022. [Google Scholar]; The document discusses common performance metrics used in machine learning classification, and introduces the expected cost (EC) metric. It compares these metrics and argues that EC is superior due to its generality, simplicity, and intuitive nature. Additionally, it highlights the potential of EC in measuring calibration and optimal decision-making using class posteriors.
- [22].Geirhos Robert, Jacobsen Jörn-Henrik, Michaelis Claudio, Zemel Richard, Brendel Wieland, Bethge Matthias, and Wichmann Felix A. Shortcut learning in deep neural networks. Nature Machine Intelligence, 2(11):665–673, November 2020. [Google Scholar]
- [23].Gooding Mark J, Smith Annamarie J, Tariq Maira, Aljabar Paul, Peressutti Devis, van der Stoep Judith, Reymen Bart, Emans Daisy, Hattu Djoya, van Loon Judith, et al. Comparative evaluation of autocontouring in clinical practice: a practical method using the turing test. Medical physics, 45(11):5105–5115, 2018. [DOI] [PubMed] [Google Scholar]
- [24].Gruber Sebastian Gregor and Buettner Florian. Better uncertainty calibration via proper scores for classification and beyond. In Advances in Neural Information Processing Systems, 2022. [Google Scholar]
- [25].Haugen Trine B, Hicks Steven A, Andersen Jorunn M, Witczak Oliwia, Hammer Hugo L, Borgli Rune, Halvorsen Pål, and Riegler Michael. Visem: A multimodal video dataset of human spermatozoa. In Proceedings of the 10th ACM Multimedia Systems Conference, pages 261–266, 2019. [Google Scholar]
- [26].Honauer Katrin, Maier-Hein Lena, and Kondermann Daniel. The hci stereo metrics: Geometry-aware performance analysis of stereo algorithms. In Proceedings of the IEEE International Conference on Computer Vision, pages 2120–2128, 2015. [Google Scholar]
- [27].Ibrahim Hussein, Liu Xiaoxuan, Zariffa Nevine, Morris Andrew D, and Denniston Alastair K. Health data poverty: an assailable barrier to equitable digital health care. Lancet Digit Health, 3(4):e260–e265, April 2021. [DOI] [PubMed] [Google Scholar]
- [28].Jaeger Paul F, Lüth Carsten T, Klein Lukas, and Bungert Till J. A call to reflect on evaluation practices for failure detection in image classification. International Conference on Learning Representations, 2023. [Google Scholar]
- [29].Jäger Paul Ferdinand. Challenges and opportunities of end-to-end learning in medical image classification. Karlsruher Institut für Technologie, 2020. [Google Scholar]
- [30].Jannin Pierre. Towards responsible research in digital technology for health care. arXiv, September 2021. [Google Scholar]
- [31].Kang Feng, Jin Rong, and Sukthankar Rahul. Correlated label propagation with application to multi-label learning. In 2006 IEEE Computer Society Conference on Computer Vision and Pattern Recognition (CVPR’06), volume 2, pages 1719–1726. IEEE, 2006. [Google Scholar]
- [32].Kelly Christopher J, Karthikesalingam Alan, Suleyman Mustafa, Corrado Greg, and King Dominic. Key challenges for delivering clinical impact with artificial intelligence. BMC medicine, 17:1–9, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Khan Daanish Ali, Li Linhong, Sha Ninghao, Liu Zhuoran, Jimenez Abelino, Raj Bhiksha, and Singh Rita. Non-determinism in neural networks for adversarial robustness. arXiv preprint arXiv:1905.10906, 2019. [Google Scholar]
- [34].Kirillov Alexander, He Kaiming, Girshick Ross, Rother Carsten, and Dollár Piotr. Panoptic segmentation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pages 9404–9413, 2019. [Google Scholar]
- [35].Kofler Florian, Ezhov Ivan, Isensee Fabian, Berger Christoph, Korner Maximilian, Paetzold Johannes, Li Hongwei, Shit Suprosanna, McKinley Richard, Bakas Spyridon, et al. Are we using appropriate segmentation metrics? Identifying correlates of human expert perception for CNN training beyond rolling the DICE coefficient. arXiv preprint arXiv:2103.06205v1, 2021. [Google Scholar]
- [36].Kofler Florian, Shit Suprosanna, Ezhov Ivan, Fidon Lucas, Al-Maskari Rami, Li Hongwei, Bhatia Harsharan, Loehr Timo, Piraud Marie, Erturk Ali, et al. blob loss: instance imbalance aware loss functions for semantic segmentation. arXiv preprint arXiv:2205.08209, 2022. [Google Scholar]
- [37].Konukoglu Ender, Glocker Ben, Ye Dong Hye, Criminisi Antonio, and Pohl Kilian M. Discriminative segmentation-based evaluation through shape dissimilarity. IEEE transactions on medical imaging, 31(12):2278–2289, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Kottner Jan, Audigé Laurent, Brorson Stig, Donner Allan, Gajewski Byron J, Hróbjartsson Asbjørn, Roberts Chris, Shoukri Mohamed, and Streiner David L. Guidelines for reporting reliability and agreement studies (grras) were proposed. International journal of nursing studies, 48(6):661–671, 2011. [DOI] [PubMed] [Google Scholar]
- [39].Lacoste Alexandre, Luccioni Alexandra, Schmidt Victor, and Dandres Thomas. Quantifying the carbon emissions of machine learning. arXiv preprint arXiv:1910.09700, 2019. [Google Scholar]
- [40].Lannelongue Loïc, Grealey Jason, and Inouye Michael. Green algorithms: quantifying the carbon footprint of computation. Advanced science, 8(12):2100707, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Lavin Alexander, Gilligan-Lee Ciarán M, Visnjic Alessya, Ganju Siddha, Newman Dava, Ganguly Sujoy, Lange Danny, Baydin Atílím Güneş, Sharma Amit, Gibson Adam, et al. Technology readiness levels for machine learning systems. Nature Communications, 13(1):1–19, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].van Leeuwen David A and Brümmer Niko. An introduction to application-independent evaluation of speaker recognition systems. In Speaker classification I, pages 330–353. Springer, 2007. [Google Scholar]
- [43].Lennerz Jochen K, Green Ursula, Williamson Drew FK, and Mahmood Faisal. A unifying force for the realization of medical ai. npj Digital Medicine, 5(1):1–3, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Liang Kung-Yee and Zeger Scott L. Longitudinal data analysis using generalized linear models. Biometrika, 73(1):13–22, 1986. [Google Scholar]
- [45].Liu Xiaoqi, Parks Kelsey, Saknite Inga, Reasat Tahsin, Cronin Austin D, Wheless Lee E, Dawant Benoit M, and Tkaczyk Eric R. Baseline photos and confident annotation improve automated detection of cutaneous graft-versus-host disease. Clinical hematology international, 3(3):108, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Ljosa Vebjorn, Sokolnicki Katherine L, and Carpenter Anne E. Annotated high-throughput microscopy image sets for validation. Nature methods, 9(7):637–637, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Maier-Hein Lena, Eisenmann Matthias, Reinke Annika, Onogur Sinan, Stankovic Marko, Scholz Patrick, Arbel Tal, Bogunovic Hrvoje, Bradley Andrew P, Carass Aaron, et al. Why rankings of biomedical image analysis competitions should be interpreted with care. Nature communications, 9(1):1–13, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]; With this comprehensive analysis of biomedical image analysis competitions (challenges), the authors initiated a shift in how such challenges are designed, performed, and reported in the biomedical domain. Its concepts and guidelines have been adopted by reputed organizations such as MICCAI.
- [48].Maier-Hein Lena, Wagner Martin, Ross Tobias, Reinke Annika, Bodenstedt Sebastian, Full Peter M, Hempe Hellena, Mindroc-Filimon Diana, Scholz Patrick, Tran Thuy Nuong, et al. Heidelberg colorectal data set for surgical data science in the sensor operating room. Scientific data, 8(1):1–11, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Maier-Hein Lena, Reinke Annika, Christodoulou Evangelia, Glocker Ben, Godau Patrick, Isensee Fabian, Kleesiek Jens, Kozubek Michal, Reyes Mauricio, Riegler Michael A, et al. Metrics reloaded: Pitfalls and recommendations for image analysis validation. arXiv preprint arXiv:2206.01653, 2022. [Google Scholar]
- [50].Mais Lisa, Hirsch Peter, and Kainmueller Dagmar. Patchperpix for instance segmentation. In European Conference on Computer Vision, pages 288–304. Springer, 2020. [Google Scholar]
- [51].Margolin Ran, Zelnik-Manor Lihi, and Tal Ayellet. How to evaluate foreground maps? In Proceedings of the IEEE conference on computer vision and pattern recognition, pages 248–255, 2014. [Google Scholar]
- [52].McCradden Melissa D, Anderson James A, Stephenson Elizabeth A, Drysdale Erik, Erdman Lauren, Goldenberg Anna, and Shaul Randi Zlotnik. A research ethics framework for the clinical translation of healthcare machine learning. Am. J. Bioeth, pages 1–15, January 2022. [DOI] [PubMed] [Google Scholar]
- [53].Meilă Marina. Comparing clusterings by the variation of information. In Learning theory and kernel machines, pages 173–187. Springer, 2003. [Google Scholar]
- [54].Meissner G, Nern A, Dorman Z, DePasquale GM, Forster K, Gibney T, Hausenfluck JH, He Y, Iyer N, Jeter J, et al. A searchable image resource of drosophila gal4-driver expression patterns with single neuron resolution. BioRxiv, page 2020.05.29.080473, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Moons Karel GM, Altman Douglas G, Reitsma Johannes B, Ioannidis John PA, Macaskill Petra, Steyerberg Ewout W, Vickers Andrew J, Ransohoff David F, and Collins Gary S. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (tripod): explanation and elaboration. Annals of internal medicine, 162(1):W1–W73, 2015. [DOI] [PubMed] [Google Scholar]
- [56].Nagao Yukiko, Sakamoto Mika, Chinen Takumi, Okada Yasushi, and Takao Daisuke. Robust classification of cell cycle phase and biological feature extraction by image-based deep learning. Molecular biology of the cell, 31(13):1346–1354, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Nasa Prashant, Jain Ravi, and Juneja Deven. Delphi methodology in healthcare research: how to decide its appropriateness. World Journal of Methodology, 11(4):116, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Oakden-Rayner Luke, Dunnmon Jared, Carneiro Gustavo, and Ré Christopher. Hidden stratification causes clinically meaningful failures in machine learning for medical imaging. Proc ACM Conf Health Inference Learn (2020), 2020: 151–159, April 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Obermeyer Ziad, Powers Brian, Vogeli Christine, and Mullainathan Sendhil. Dissecting racial bias in an algorithm used to manage the health of populations. Science, 366(6464):447–453, October 2019. [DOI] [PubMed] [Google Scholar]
- [60].Park Seong Ho, Han Kyunghwa, Jang Hye Young, Park Ji Eun, Lee June-Goo, Kim Dong Wook, and Choi Jaesoon. Methods for Clinical Evaluation of Artificial Intelligence Algorithms for Medical Diagnosis. Radiology, 306(1):20–31, January 2023. ISSN 0033–8419. doi: 10.1148/radiol.220182. URL https://pubs.rsna.org/doi/10.1148/radiol.220182. Publisher: Radiological Society of North America. [DOI] [PubMed] [Google Scholar]
- [61].Patterson David, Gonzalez Joseph, Le Quoc, Liang Chen, Munguia Lluis-Miquel, Rothchild Daniel, So David, Texier Maud, and Dean Jeff. Carbon emissions and large neural network training. arXiv, April 2021. [Google Scholar]
- [62].Perez-Lebel Alexandre, Le Morvan Marine, and Varoquaux Gaël. Beyond calibration: estimating the grouping loss of modern neural networks. International Conference on Learning Representations, 2023. [Google Scholar]
- [63].Rand William M. Objective criteria for the evaluation of clustering methods. Journal of the American Statistical association, 66(336):846–850, 1971. [Google Scholar]
- [64].Reinke Annika, Eisenmann Matthias, Onogur Sinan, Stankovic Marko, Scholz Patrick, Full Peter M, Bogunovic Hrvoje, Landman Bennett A, Maier Oskar, Menze Bjoern, et al. How to exploit weaknesses in biomedical challenge design and organization. In International Conference on Medical Image Computing and Computer-Assisted Intervention, pages 388–395. Springer, 2018. [Google Scholar]
- [65].Reinke Annika, Eisenmann Matthias, Tizabi Minu D, Sudre Carole H, Rädsch Tim, Antonelli Michela, Arbel Tal, Bakas Spyridon, Cardoso M Jorge, Cheplygina Veronika, et al. Common limitations of image processing metrics: A picture story. arXiv preprint arXiv:2104.05642, 2021. [Google Scholar]
- [66].Reinke Annika, Tizabi Minu D., Baumgartner Michael, Eisenmann Matthias, Heckmann-Nötzel Doreen, Kavur Emre, Rädsch Tim, Sudre Carole, et al. Understanding metric-related pitfalls in image analysis validation. arXiv preprint arXiv:2302.01790; sister publication jointly submitted with this work, 2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Riley Richard D, Ensor Joie, Snell Kym IE, Debray Thomas PA, Altman Doug G, Moons Karel GM, and Collins Gary S. External validation of clinical prediction models using big datasets from e-health records or ipd meta-analysis: opportunities and challenges. bmj, 353, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Roß Tobias, Bruno Pierangela, Reinke Annika, Wiesenfarth Manuel, Koeppel Lisa, Full Peter M, Pekdemir Bünyamin, Godau Patrick, Trofimova Darya, Isensee Fabian, et al. How can we learn (more) from challenges? a statistical approach to driving future algorithm development. arXiv preprint arXiv:2106.09302, 2021. [Google Scholar]
- [69].Sage Daniel, Kirshner Hagai, Pengo Thomas, Stuurman Nico, Min Junhong, Manley Suliana, and Unser Michael. Quantitative evaluation of software packages for single-molecule localization microscopy. Nature methods, 12(8): 717–724, 2015. [DOI] [PubMed] [Google Scholar]
- [70].Schulam Peter and Saria Suchi. Can you trust this prediction? auditing pointwise reliability after learning. In Chaudhuri Kamalika and Sugiyama Masashi, editors, Proceedings of the Twenty-Second International Conference on Artificial Intelligence and Statistics, volume 89 of Proceedings of Machine Learning Research, pages 1022–1031. PMLR, 2019. [Google Scholar]
- [71].Schulz Kenneth F, Altman Douglas G, Moher David, and CONSORT Group*. Consort 2010 statement: updated guidelines for reporting parallel group randomized trials. Annals of internal medicine, 152(11):726–732, 2010. [DOI] [PubMed] [Google Scholar]
- [72].Shah Nigam H, Milstein Arnold, and Bagley Steven C. Making machine learning models clinically useful. Jama, 322 (14):1351–1352, 2019. [DOI] [PubMed] [Google Scholar]
- [73].Simpson Amber L, Antonelli Michela, Bakas Spyridon, Bilello Michel, Farahani Keyvan, Van Ginneken Bram, Kopp-Schneider Annette, Landman Bennett A, Litjens Geert, Menze Bjoern, et al. A large annotated medical image dataset for the development and evaluation of segmentation algorithms. arXiv preprint arXiv:1902.09063, 2019. [Google Scholar]
- [74].Sounderajah Viknesh, Ashrafian Hutan, Aggarwal Ravi, De Fauw Jeffrey, Denniston Alastair K, Greaves Felix, Karthikesalingam Alan, King Dominic, Liu Xiaoxuan, Markar Sheraz R, McInnes Matthew D F, Panch Trishan, Pearson-Stuttard Jonathan, Ting Daniel S W, Golub Robert M, Moher David, Bossuyt Patrick M, and Darzi Ara. Developing specific reporting guidelines for diagnostic accuracy studies assessing AI interventions: The STARD-AI steering group. Nat. Med, 26(6):807–808, June 2020. [DOI] [PubMed] [Google Scholar]
- [75].Steyerberg Ewout W, Vickers Andrew J, Cook Nancy R, Gerds Thomas, Gonen Mithat, Obuchowski Nancy, Pencina Michael J, and Kattan Michael W. Assessing the performance of prediction models: a framework for some traditional and novel measures. Epidemiology (Cambridge, Mass.), 21(1):128, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Strubell Emma, Ganesh Ananya, and McCallum Andrew. Energy and policy considerations for deep learning in NLP. arXiv, June 2019. [Google Scholar]
- [77].Summers Cecilia and Dinneen Michael J. Nondeterminism and instability in neural network optimization. In International Conference on Machine Learning, pages 9913–9922. PMLR, 2021. [Google Scholar]
- [78].Taha Abdel Aziz and Hanbury Allan. Metrics for evaluating 3d medical image segmentation: analysis, selection, and tool. BMC medical imaging, 15(1):1–28, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]; The paper discusses the importance of effective metrics for evaluating the accuracy of 3D medical image segmentation algorithms. The authors analyze existing metrics, propose a selection methodology, and develop a tool to aid researchers in choosing appropriate evaluation metrics based on the specific characteristics of the segmentation task.
- [79].Targosz Anna, Przystałka Piotr, Wiaderkiewicz Ryszard, and Mrugacz Grzegorz. Semantic segmentation of human oocyte images using deep neural networks. BioMedical Engineering OnLine, 20(1):40, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].The Institute for Ethical Ai and Machine Learning. The institute for ethical AI & machine learning. https://ethical.institute/principles.html, 2018. Accessed: 2022-5-21. [Google Scholar]
- [81].Tirian Laszlo and Dickson Barry J. The vt gal4, lexa, and split-gal4 driver line collections for targeted expression in the drosophila nervous system. BioRxiv, page 198648, 2017. [Google Scholar]
- [82].Tran Thuy N, Adler Tim, Yamlahi Amine, Christodoulou Evangelia, Godau Patrick, Reinke Annika, Tizabi Minu D, Sauer Peter, Persicke Tillmann, Albert Jörg G., and Maier-Hein Lena. Sources of performance variability in deep learning-based polyp detection. arXiv preprint arXiv:2211.09708, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Ulman Vladimír, Maška Martin, Magnusson Klas EG, Ronneberger Olaf, Haubold Carsten, Harder Nathalie, Matula Pavel, Matula Petr, Svoboda David, Radojevic Miroslav, et al. An objective comparison of cell-tracking algorithms. Nature methods, 14(12):1141–1152, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Usatine Richard and Manci Rachel. Dermoscopedia, 2021. https://dermoscopedia.org/File:DF_chinese_dms.JPG.
- [85].Vaassen Femke, Hazelaar Colien, Vaniqui Ana, Gooding Mark, van der Heyden Brent, Canters Richard, and van Elmpt Wouter. Evaluation of measures for assessing time-saving of automatic organ-at-risk segmentation in radiotherapy. Physics and Imaging in Radiation Oncology, 13:1–6, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Van Hoorde Kirsten, Van Huffel Sabine, Timmerman Dirk, Bourne Tom, and Van Calster Ben. A spline-based tool to assess and visualize the calibration of multiclass risk predictions. Journal of biomedical informatics, 54:283–293, 2015. [DOI] [PubMed] [Google Scholar]
- [87].Vickers Andrew J, Van Calster Ben, and Steyerberg Ewout W. Net benefit approaches to the evaluation of prediction models, molecular markers, and diagnostic tests. bmj, 352, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Wiesenfarth Manuel, Reinke Annika, Landman Bennett A, Eisenmann Matthias, Saiz Laura Aguilera, Cardoso M Jorge, Maier-Hein Lena, and Kopp-Schneider Annette. Methods and open-source toolkit for analyzing and visualizing challenge results. Scientific reports, 11(1):1–15, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Anthony Lasse F Wolff, Kanding Benjamin, and Selvan Raghavendra. Carbontracker: Tracking and predicting the carbon footprint of training deep learning models. arXiv, July 2020. [Google Scholar]
- [90].Zhang Ying, Xie Yubin, Liu Wenzhong, Deng Wankun, Peng Di, Wang Chenwei, Xu Haodong, Ruan Chen, Deng Yongjie, Guo Yaping, et al. Deepphagy: a deep learning framework for quantitatively measuring autophagy activity in saccharomyces cerevisiae. Autophagy, 16(4):626–640, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No data was used in this study.


