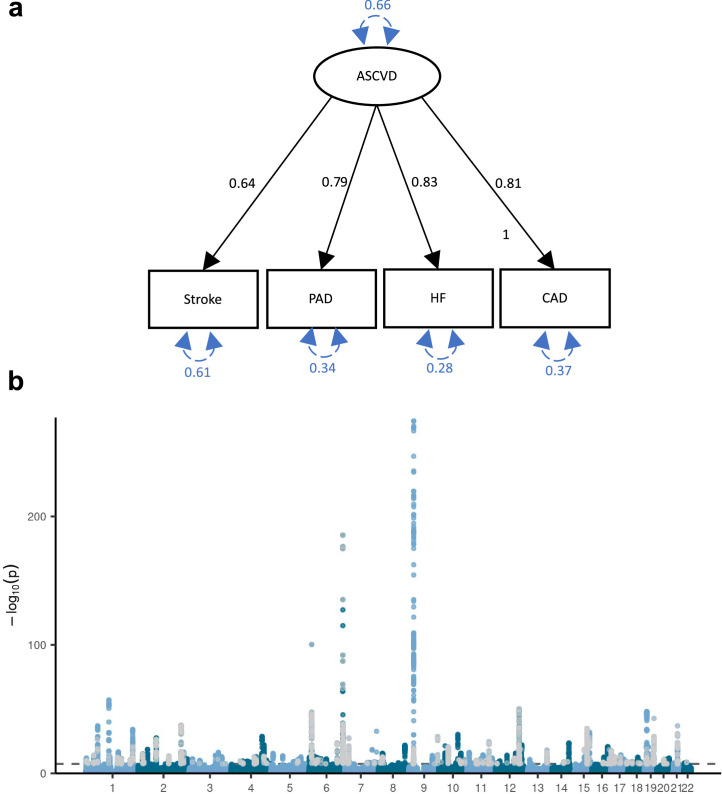
Extended Data Fig. 3. Genetic liability to ASCVD latent factor.
a, Latent atherosclerotic CVD (ASCVD) model, defined by stroke, peripheral artery disease (PAD), heart failure (HF), and coronary artery disease (CAD). All ‘observed’ traits are based on GWAS summary statistics. Results are from confirmatory factor analysis in Genomic SEM, and standardized factor loadings are given for each path. Circular dashed arrows give the trait variance. b, Manhattan plot of the GWAS on ASCVD, with each dot representing a SNP with its position on the x-axis and its P-value on the y-axis. Genome-wide significant SNPs with a significant heterogeneity Q (with a strong effect on one or some of the indicators that was not well explained through the common latent factor) are displayed in grey. The dashed line indicates the genome-wide significance threshold (P < 5e-8). P-values were computed using a two-sided Z-test. a-b Sample sizes for underlying GWAS summary statistics are reported in Supplementary Table 1. AF=Atrial Fibrillation; CAD=Coronary Artery Disease; HF=Heart Failure; MDD=Major depressive disorder; PAD=Peripheral Artery Disease.