Figure 3.
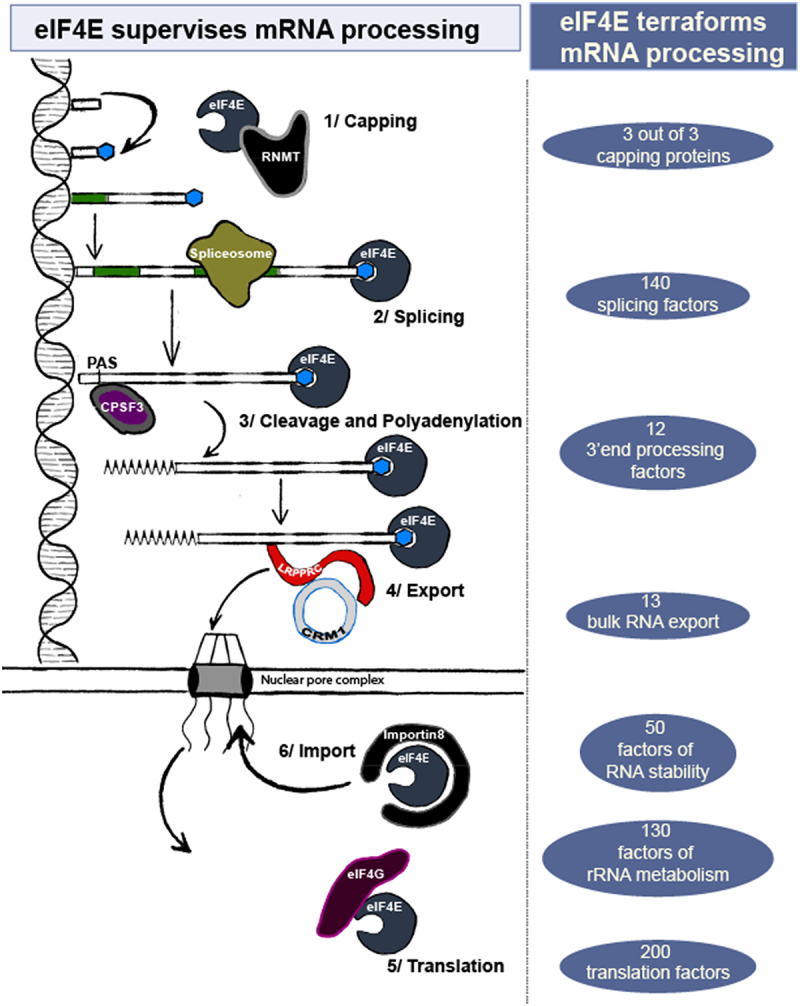
eIF4E can directly influence RNA fate or indirectly through its capacity to terraform the RNA processing landscape. On the left, interactions between eIF4E target mRNAs and the noted machineries are shown. On the right, numbers of mRNAs in a given process were identified in nuclear, endogenous eIF4E RIP-Seq [54] GSE63265_LY1_4EIP_allreps_counts.Txt.gz and/or RNA-seq splicing data segregated on high and normal-eIF4E AML specimens [100] (https://leucegene.ca/). These strongly suggest that eIF4E can influence a broad array of factors responsible for RNA processing thereby terraforming the RNA processing landscape. Functional categories were assigned using Metascape (metascape.Org).
