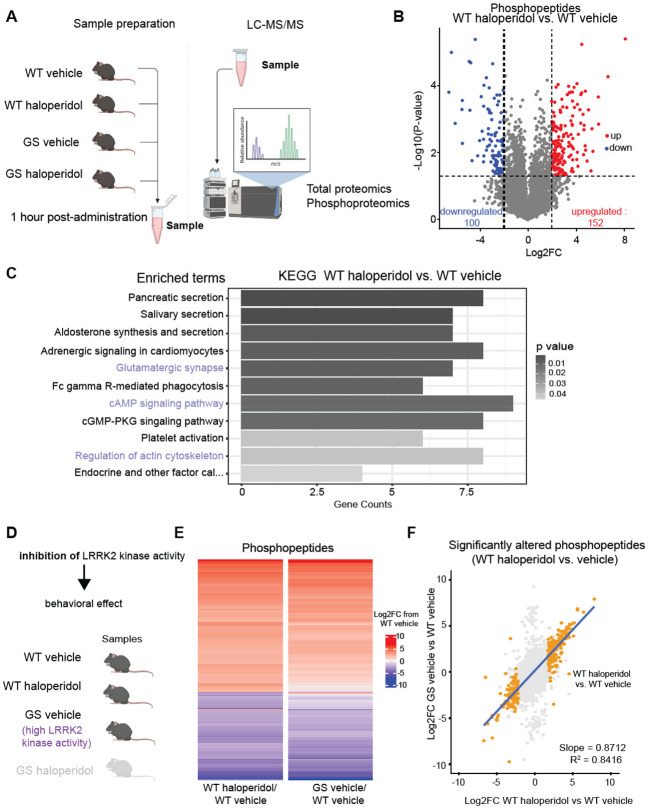
Figure 3: Haloperidol treatment induces similar changes in the striatal phosphoproteome as increased LRRK2 kinase activity.
A. Schematic of experimental procedures. WT and GS mutant mice were treated with haloperidol or vehicle, and striatal samples were harvested after 1 hour. LC-MS/MS analyses for total proteins and phosphopeptides were conducted on the same striatal samples. N=3 mice/group. Parts of the schematic were created with BioRender.com.
B. Volcano plot comparing the phosphopeptides between haloperidol and vehicle-treated WT mice. Phosphopeptides that are significantly differentially regulated (∣Log2FC∣ > 2 and unadjusted p-value ≤ 0.05 by multiple unpaired t-tests) are colored red and blue for up- and down-regulated, respectively.
C. Results of Gene Set Enrichment Analysis (GSEA) of the KEGG gene set for genes with at least one differentially regulated phosphopeptide in WT haloperidol vs vehicle-treated comparison. Pathways displayed are significantly differently regulated (adjusted p-value ≤ 0.05 by Fisher's test). The length of bars reflects the number of genes in the pathway whose phosphostate is differentially regulated; bars are shaded by adjusted p-value. Highlighted pathways are related to established LRRK2 functions.
D. Diagram of genetic and pharmacological conditions for groups analyzed in E. Created with BioRender.com.
E. Heatmap of effect size (Log2FC) of either haloperidol treatment or LRRK2-GS for all differentially abundant phosphopeptides in the WT haloperidol-treated vs vehicle-treated comparison (∣Log2FC∣ > 2 and unadjusted p-value ≤ 0.05 by multiple unpaired t-tests). Each bar represents a phosphopeptide. Color reflects Log2FC difference from WT saline condition.
F. Correlation plot for E, comparing GS vehicle/WT vehicle to WT haloperidol-vehicle effect size. All detected phosphopeptides were mapped, and phosphopeptides significantly altered between WT haloperidol vs. vehicle are highlighted. Only highlighted values are used for correlation calculation. Blue line represents the line of best fit.