Figure 8. Env regions targeted by the A32/17b/246D/CJF-III-288 cocktail.
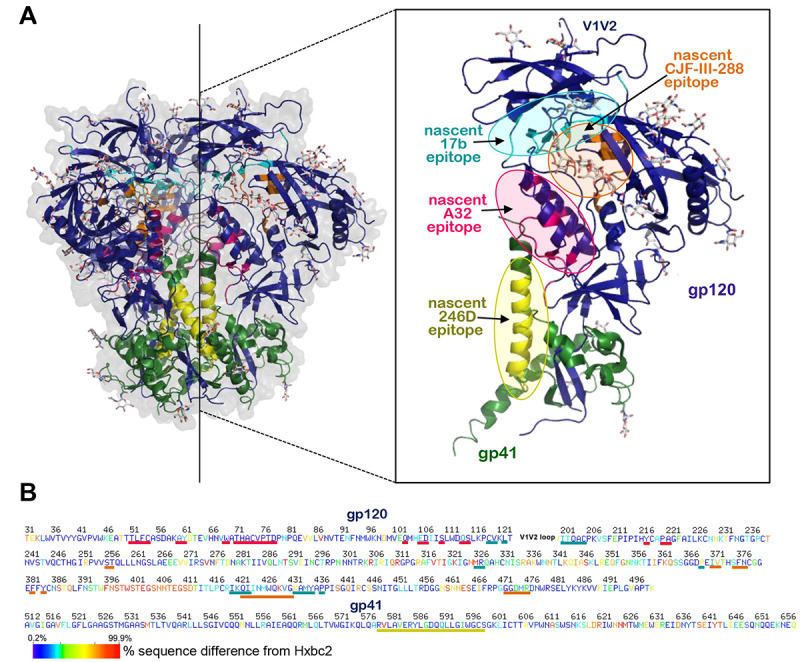
(A) Location of A32 (anti-cluster A), 17b (anti-CoRBS), 246D (anti-gp41 cluster I) and CJF-III-288 (CD4mc) binding sites within the unliganded HIV-1 trimer. The BG505 SOSIP.664 gp140 unliganded trimer (PDB code: 4ZMJ) is shown, colored green and dark blue for gp41 and gp120 protomers. The nascent epitopes for A32, 17b, 246D and CJF-III-288 are colored magenta, cyan, yellow and orange, respectively. The nascent epitopes are defined as gp120 contact residues contributing buried surface area (BSA) to the Env antigen-Fab interface calculated from available structures: A32 (PDB code: 4YC2), 17b (PDB code: 6CM3) and CJF-III-288 (PDB code:8FM3). Epitope for 246D is as defined by peptide scanning in (48). The blow-up view shows insight into one gp41-gp120 protomer with epitopes. (B) Sequence conservation of A32, 17b, 246D and CJF-III-288 binding regions among all listed Env sequences from the Los Alamos Data Base. The sequence of gp120 and gp41, excluding the V1V2 region, is shown based on the conservation of each residue e.g. residues differed 0.2–7% and 87–99.9% from the Hxbc2 sequence colored with dark blue and red. Lines (A32: magenta, 17b: cyan, 246D: yellow and CJF-III-288: orange) below the sequence indicate the epitope footprints.
