Abstract
Biological adaptive systems share some common features: variation among their constituent elements and continuity of core information. Some of them, such as the immune system, are endowed with memory of past events. In this study we provide direct evidence that evolving viral quasispecies possess a molecular memory in the form of minority components that populate their mutant spectra. The experiments have involved foot-and-mouth disease virus populations with known evolutionary histories. The composition and behavior of the viral population in response to a selective constraint were influenced by past evolutionary history in a way that could not be predicted from examination of consensus nucleotide sequences of the viral populations. The molecular memory of the viral quasispecies influenced both the nature and the intensity of the response of the virus to a selective constraint.
RNA viruses are the most widespread group of molecular parasites and the etiological agents of many important human, animal, and plant diseases such as AIDS and several forms of hepatitis or hemorrhagic fevers, among many others (11, 29). During their replication, RNA genomes mutate at average rates of 10−3 to 10−5 substitutions per nucleotide copied (3, 8, 12). RNA virus populations consist of complex and dynamic mutant distributions termed viral quasispecies (13–16). The quasispecies concept was first proposed by Eigen, Schuster, and Biebricher (13–16) to describe error-prone replication and self-organization of primitive macromolecules thought to carry information as precursors of more complex life forms (14). The concept has found an extremely valuable application in the understanding of present-day RNA viruses and other RNA genetic elements, which are subjected to a continuous process of genetic change, competition, and selection of the most fit mutant distributions (3, 8, 10, 11, 14, 15, 22).
A viral quasispecies is defined by a master sequence and a mutant spectrum. The master sequence is the dominant nucleotide sequence in the genomic distribution. It often shows the highest selection value among the mutants present, and it may or may not coincide with the consensus or average sequence of the distribution. The mutant spectrum is the ensemble of error copies rated according to their relative replication abilities in the context of the ensemble of mutants (10, 14–16, 22). The quasispecies as a whole, rather than its individual components, acts as a unit of selection (13–16). In the course of natural infections with RNA viruses, both the master sequences and the mutant spectra often have a very transient existence since the population equilibrium (a selective equilibrium or selective rating of all mutants that may appear) of viral genomes is frequently perturbed by environmental modifications (8, 22). The mutant spectrum is a repository of genetic and phenotypic variants with the potential to become dominant in response to environmental demands, as has been shown, for example, with antibody or cytotoxic-T-cell escape mutants and inhibitor-resistant mutants of pathogenic viruses (5, 8, 11, 22).
Mobilization of minority elements in response to an external stimulus is typical of complex adaptive systems such as the immune systems of vertebrates (2, 24, 25). In contact with a foreign antigen, cells of the immune system undergo a cascade of recombination and mutation events that lead to selection and clonal expansion of cells showing the highest affinity for the antigen. This process generates long-lived memory T cells which are able to maintain long-term immunity that may last throughout the entire lifetime of the organism (2, 24, 25). In the evolution of a viral quasispecies, the genomes hidden in the mutant spectrum may constitute a molecular record of the past evolutionary history of the population. This record would represent a molecular memory with obvious implications for the types and rapidity of the responses of the population to selective constraints. With the aim of exploring the possible existence of a molecular memory in viral quasispecies, we have designed experiments using the animal pathogen foot-and-mouth disease virus (FMDV), a representative of the Picornaviridae family (30). FMDV populations with a well-defined evolutionary history under a controlled cell culture environment have been analyzed with regard to their response to a selective constraint that had been previously experienced by the population. We have also tested the presence in the mutant spectra of the viral populations of minority components that may reveal in a specific fashion their past history. The results provide direct evidence for the existence of a molecular memory in viral quasispecies and illustrate the influence of such a memory in responses to selective constraints.
MATERIALS AND METHODS
Description of the viral populations used.
FMDV is an aphthovirus of the Picornaviridae family (30). FMDV C-S8c1 is a standard biological clone of FMDV that has been extensively used for experiments of FMDV evolution in cell culture (9, 33). FMDV RED is a monoclonal antibody (MAb)-resistant (MAR) mutant of FMDV C-S8c1 p100, a population derived from serially passaging clone C-S8c1 100 times in BHK-21 cells (2 × 106 cells infected with 4 × 106 to 8 × 106 PFU of virus per passage) (27). FMDV RED includes the amino acid replacement Gly-142→Glu at a highly conserved RGD triplet located at the exposed G-H loop of capsid protein VP1 (1, 27). This loop serves as an integrin recognition site for virus entry into the cell and also as a site of interaction with neutralizing antibodies (4, 23, 31, 34, 35).
Serial plaque-to-plaque transfers of FMDV C-S8c1 led to several highly debilitated clones due to an accentuation of Muller's ratchet effect (17). One such clone is C229. FMDV C229 p50 is the population that resulted from subjecting clone C229 to 50 serial infections in BHK-21 cells (4 × 106 cells infected with 1 × 106 to 1 × 107 PFU per passage) (18). The fitness gain of clone C229 was monitored for 100 passages (18). Viral populations analyzed in the present study, either FMDV RED or FMDV C229, originated from plaque-purified virus to ensure that viral progenies were derived from single infectious genomes.
Infections, mutant isolation, and biological cloning.
Procedures for the infection of BHK-21 cells with FMDV for the isolation of MAR mutants resistant to neutralization by MAb SD6 and for biological cloning have been previously described (27, 28, 33). MAb SD6 is a neutralizing MAb raised against FMDV C-S8c1 which recognizes an epitope located within the G-H loop of capsid protein VP1 (28). Relative fitness values were calculated as previously described (18, 21). To control for the absence of contamination with other FMDVs, parallel passages of the supernatants of mock-infected cells were carried out throughout the experiments, with no signs of cytopathology or infectivity in the cultures. Contamination was also ruled out by nucleotide sequencing of genomic regions which included diagnostic mutations for the viruses under study. Specifically, the capsid of all FMDV RED revertants included mutations characteristic of this lineage and absent in the standard FMDV C-S8c1 (27), and the clones derived from C229 p50 included a point deletion at genomic residue 1056, which is specific to the C229 lineage (17, 18).
Viral RNA extraction, RT-PCR amplification, and nucleotide sequencing.
Viral RNA was extracted as previously described (17). Amplification of genomic residues 569 to 1200 and 3550 to 3750 by reverse transcriptase PCR (RT-PCR) was performed using about 1% (1 ng) of viral RNA from a single plaque and avian myeloblastosis virus RT (Promega) and Ampli-Taq (Perkin-Elmer), as specified by the manufacturers. The oligonucleotides primers used to amplify the 569-to-1200 region corresponded to genomic residues 569 to 588 (sense) and were complementary to genomic positions 1200 to 1183 (antisense); primers used to amplify the 3550-to-3750 region corresponded to genomic residues 2817 to 2838 (sense) and were complementary to genomic positions 3896 to 3873 (antisense). Residue numbering is according to the work of Escarmís et al. (17). DNA sequencing was done with an ABI 373 automatic sequencer.
Molecular cloning of FMDV genomes.
Molecular cloning was performed by subjecting about 7 ng of viral RNA to RT-PCR amplification using avian myeloblastosis virus RT and Pfu I DNA polymerase (Promega) (7), as specified by the manufacturer. The oligonucleotide primers were those described above for the Ampli-Taq amplification reaction and contained the SacI and BamHI restriction sites. Amplified DNAs were purified, digested with SacI and BamHI, and ligated to pGEM-4Z digested with the same restriction enzymes. Transformation into Escherichia coli DH5-α and screening for positive recombinants were carried out by following standard procedures (32).
RESULTS
Rescue of FMDV RED mutants from a revertant population.
A number of mutants lacking the RGD triplet in capsid protein VP1 were isolated as MAR mutants of a population of FMDV obtained by subjecting clone FMDV C-S8c1 to 100 serial passages in BHK-21 cells (27, 31) (Fig. 1). One of the mutants included an RED instead of an RGD triplet. The genetic stability of FMDV RED was tested by subjecting a biological clone to multiple serial passages in BHK-21 cells (Fig. 1). Nucleotide sequencing established that reversion occurred in each of the three passage series and that no FMDV RED was detectable in the consensus sequence of the populations at passage 10 (Fig. 2). From the slopes of the change of proportion of the two diagnostic sequences as a function of passage number, the fitness of FMDV RED relative to that of FMDV RGD was 0.40 ± 0.08. To probe whether the virus population could maintain a molecular memory of its origin as an RED mutant, 15 additional serial infections were carried out at a multiplicity of infection of 0.1 PFU per cell (which represented about 120 doublings of the amount of infectious particles), and the proportions of SD6 MAR mutants in the viral populations at passages 15 and 25 were determined (Table 1). MAR mutant frequencies were 25- to 1,000-fold higher than for FMDV C-S8c1 and 3- to 120-fold higher than for FMDV C-S8c1 p100, when these populations were subjected to the same selection procedure (Table 1). Furthermore, all MAR mutants included an RED triplet whereas none of 81 MAR mutants derived from FMDV C-S8c1 and only 4 out of 31 MAR mutants derived from FMDV C-S8c1 p100 included this amino acid substitution (P < 0.001; χ2 test) (Table 1).
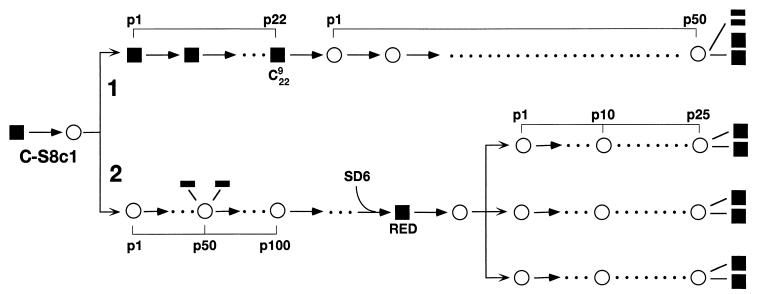
FIG. 1.
Passage history of the FMDV populations employed in the present study. Filled squares and rectangles represent biological and molecular clones, respectively; empty circles represent uncloned viral populations. In lineage 1 FMDV clone C-S8c1 was subjected to 22 serial plaque transfers to obtain clone C229 (17). Then it was subjected to 50 serial infections in BHK-21 cells, as detailed in Materials and Methods. At passage 50 the population was subjected to molecular and biological cloning and individual clones were analyzed by nucleotide sequencing. In lineage 2, FMDV clone C-S8c1 was passaged 100 times in BHK-21 cells (27, 31), as detailed in Materials and Methods. At passage 50 the population was analyzed by molecular cloning and nucleotide sequencing. At passage 100, a MAb SD6-resistant mutant was obtained (RED). Triplicate samples of this virus were serially passaged 25 times at a multiplicity of infection of 0.1 PFU per cell (4 × 106 cells infected with 4 × 105 PFU of virus). RNAs from individual biological clones from these populations were analyzed by nucleotide sequencing.
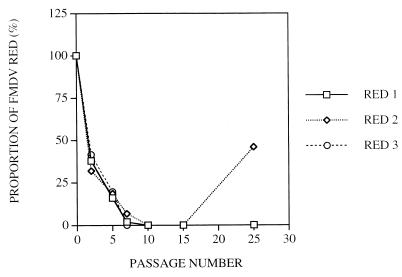
FIG. 2.
Reversion of FMDV RED. Three clonal populations of FMDV RED were serially passaged in BHK-21 cells, as detailed in Materials and Methods and in the legend to Fig. 1. At passages 0, 2, 5, 7, 10, 15, and 25 the proportions of FMDV RED and its revertant FMDV RGD were calculated from sequencing gels using a Fujifilm BAS 1500 densitometer. Fitness values (21) of FMDV RED, relative to those of the corresponding revertant viruses, were 0.43 (r2 = 0.960) for RED1, 0.46 (r2 = 0.874) for RED2, and 0.31 (r2 = 0.79) for RED3. Procedures are detailed in Materials and Methods.
TABLE 1.
Frequencies of SD6 MAR mutants in FMDV populations
| Virus populationa (culture) | MAR mutant frequencyb | No. of clones analyzedb | % of clones with RED sequence |
|---|---|---|---|
| C-S8c1 | (5.0 ± 1.6) × 10−4 | 86 | 0 |
| C-S8c1 p100 | (4.1 ± 0.5) × 10−3 | 31 | 13 |
| RGD rev. p15 (1) | 1.8 × 10−2 | 5 | 100 |
| RGD rev. p15 (2) | 1.4 × 10−2 | 5 | 100 |
| RGD rev. p15 (3) | 1.3 × 10−2 | 5 | 100 |
| RGD rev. p25 (1) | 1.6 × 10−2 | 5 | 100 |
| RGD rev. p25 (2) | 5.3 × 10−1 | 5 | 100 |
| RGD rev. p25 (3) | 2.7 × 10−2 | 5 | 100 |
Viral populations are described in Materials and Methods and in the legend to Fig. 1. “p” indicates passage number (i.e., C-S8c1 p100 means FMDV clone C-S8c1 passaged 100 times in BHK-21 cells as described in reference 27); RGD rev. viruses are the populations of FMDV RED that reverted to the RGD wild-type sequence upon passage in BHK-21 cells, as described in the legends to Fig. 1 and 2 and in Materials and Methods.
In all cases, the frequencies of MAR mutants were calculated from the number of plaques formed upon plating 2 × 102 to 1 × 105 PFU of FMDV in the presence of MAb SD6, as previously described (28). Virus from each of the plaques analyzed (86 plaques from C-S8c1, 31 plaques from C-S8c1 p100, and a total of 30 plaques from the RGD revertant populations) showed an increased resistance to neutralization by SD6 and one amino acid substitution within the G-H loop of capsid protein VP1.
To test whether bottleneck events could eliminate the molecular memory in the quasispecies, the three FMDV RED populations at passage 15 were further passaged 10 times at a multiplicity of infection of 10−4 to 10−5 PFU per cell (106 cells infected with 10 to 100 PFU per passage). SD6 MAR mutant frequencies were (1.9 ± 0.5) × 10−4. Out of 16 mutants analyzed, none included the RED sequence; instead, other amino acids changes were found at the G-H loop of VP1 (Asp 143→Gly, six clones; Leu 144→Ser, five clones; and His 146→Pro, Gly 142→Arg, Ser 139→Ile, Ser 139→Arg, and Ser 139→Gly, one clone each) (P < 0.001; χ2 test). Therefore, bottleneck events eliminated the preference to rescue MAR mutants with the RED triplet at the G-H loop of capsid protein VP1. To further exclude the possibility that the genomic sequence of RGD-containing revertants, rather than the mutant spectrum of the quasispecies, dictated selection of RED mutants by MAb SD6, the same selection by SD6 was applied to four RGD-containing clones isolated from the FMDV RED population at passage 25. Out of 12 MAR mutants analyzed, 2 included the RED sequence and 10 included different amino acid substitutions at the G-H loop of VP1 (Leu-144→Ser, 3 clones; Asp-143→Gly, 2 clones; Ser-139→Asn, 2 clones, one of which had the additional substitution Ala-138→Asn; Ala-138→Asp, 2 clones; and Gly-142→Arg, 1 clone). Again, this distribution of MAR mutants is significantly different than the one reflected in complete dominance of MAR mutants with RED obtained from RGD revertant passage 15 and passage 25 populations (data in Table 1; P<0.001; χ2 test). Therefore, memory of past history as RED-containing virus in RGD revertant passage 15 and passage 25 populations is a property of the viral quasispecies as a whole and not of the individual genomes that compose it.
Maintenance of highly altered genomic sequences dependent on passage history.
C229 is a highly debilitated clone derived from serial plaque transfers of FMDV C-S8c1 (17). A salient feature of this clone is the presence of an elongated internal polyadenylate between genomic positions 1119 and 1123 (the wild-type sequence includes four adenylates preceding the second functional AUG translation initiation codon). This polyadenylate tract is heterogeneous in length, with an average of 23 additional adenylate residues (17, 18). In the process of fitness recovery of C229 upon serial passage in BHK-21 cells, the first modification detectable in the entire genome was the reversion of this internal polyadenylate to the wild-type sequence (18). By passage 20 no detectable elongated internal polyadenylate was present in the consensus sequence of the population, and such an absence was maintained in successive passages (18). To test whether the virus population kept a molecular memory of its origin as a clone with an internal polyadenylate, the C229 population at passage 50 was subjected to molecular and biological cloning and the clones were analyzed by nucleotide sequencing (Table 2). In 6 out of 35 molecular clones, and in 4 out of 35 biological clones, genomes with a larger number of adenylate residues between genomic positions 1119 and 1123 were present. In contrast, no genomes with an increased number of adenylates at this genomic region were found in any of 40 molecular clones derived from passage 50 of a population that had C-S8c1 rather than C229 as its initial genome (0.01 > P > 0.0025; χ2 test) (Table 2). The results document the existence of a molecular memory in viral quasispecies which is imprinted in their mutant spectra as a reflection of their past evolutionary history.
TABLE 2.
Presence of additional adenylate residues between genomic positions 1119 and 1123 of FMDV populationsa
| No. of additional adenylate residues | No. of FMDV C229 p50
|
No. of FMDV C-S8c1 p50 molecular clones | |
|---|---|---|---|
| Molecular clones | Biological clones | ||
| 0 | 29 | 31 | 40 |
| 1 | 1 | 3 | 0 |
| 3 | 1 | 0 | 0 |
| 4 | 1 | 0 | 0 |
| 5 | 1 | 0 | 0 |
| 6 | 1 | 0 | 0 |
| 10 | 1 | 0 | 0 |
| 3–6b | 0 | 1 | 0 |
FMDV C229 p50 and FMDV C-S8c1 p50 are the populations obtained after passaging clones C229 and C-S8c1, respectively, in BHK-21 cells, as detailed in Materials and Methods and the legend to Fig. 1. Thirty-five and 40 molecular clones were analyzed from populations of C229 p50 and C-S8c1 p50, respectively; in addition, 35 biological clones were analyzed from populations of C229 p50. Procedures for molecular and biological cloning and nucleotide sequence determination are described in Materials and Methods.
This biological clone, derived from FMDV C229 p50 showed heterogeneity with regard to the number of internal adenylate residues between positions 1119 and 1123 of the FMDV genome (17). The figures given represent the numbers of additional adenylate residues estimated from the peak pattern in a sequencing gel.
DISCUSSION
Viral quasispecies represent complex adaptive systems (19, 20). The behavior of such systems is under intense investigations in disparate fields of research, including adaptive immunity, neural development and learning, and design of intelligent computer systems (19, 20). Common to complex adaptive systems is that they vary, yet they have continuity in the form of some heritable (or built-in) component and they are endowed with mechanisms to respond to external stimuli (19, 20). The immune systems of vertebrates exploit a memory in the form of clonal expansions of cell subsets in response to stimuli by antigens that had been previously experienced by the system (2, 19, 24, 25).
In the present report we have provided direct evidence that viral quasispecies are endowed with a molecular memory contained within the components of the mutant spectra. Batschelet et al. (3) estimated the proportions of two competing bacteriophages (W, wild type, and M, mutant) that differed in a point mutation, one becoming dominant over the other as a result of reversion and the competitive advantage of the revertant. The model formulated by Batschelet et al. (3) to describe the change in the compositions of the populations from infectious cycle N to N + 1 is
 |
1 |
 |
in which kw and km are the growth factors of the revertant (W) and mutant (M) viruses, respectively, and μw and μm are the mutation rates (in substitutions per nucleotide copied) from the wild type to the mutant and from the mutant to the wild type (reversion), respectively. This treatment can be applied to the reversion of FMDV RED (M) to FMDV RGD (W). The proportion (pN) of the revertant (W) over that of the mutant (M) at passage N is
 |
2 |
In equation 2, q is μ/ɛ, and ɛ is kw minus μw minus km plus μm (3). Assuming equal rates of mutation for FMDV RED reverting to RGD and for FMDV RGD generating RED (μ = μw = μm), the proportion of FMDV RED (pN) for a km/kw of 0.4 (the value found experimentally [Fig. 2]) can be calculated as a function of the mutation rate and passage number (3). With μ equal to 10−4, pN values for passages 10, 15, and 25 and for a very large number of passages (N ≈ ∞) are 3.7 × 10−1, 6.6 × 10−3, 1.7 × 10−4, and 1.7 × 10−4, respectively. For a μ of 10−3, the pN values are 6.3 × 10−2, 2.4 × 10−3, 1.7 × 10−3, and 1.7 × 10−3, respectively. These predicted proportions of RED mutants are lower than the proportions of MAR mutants determined experimentally (compare with values in Table 1). The discrepancy may be due to a number of assumptions that affect the values entered in equation 2: identical values for μw and μm, invariance of km/kw values in the course of passaging, etc. However, the treatment of Batschelet et al. (3), in agreement with theoretical quasispecies concepts (13–16), predicts that a population equilibrium will be reached, at which the proportion of a minority mutant will depend on its fitness relative to that of the wild-type competitor and on the mutation rate (3). This prediction provides theoretical support for the existence of a molecular memory in viral quasispecies, as documented experimentally by our results. In reality, the situation with two competing viruses is more complex than that proposed by our model, since upon replication, mutations that may alter the relative levels of fitness of the two competing viruses may occur anywhere in the genomes. As quantified in series 2 of the FMDV RED reversion experiment, an increase in the proportion of mutant virus was detected at passage 25 (Fig. 2 and Table 1), and in this case the past evolutionary history was responsible for the high frequency of a mutant with an unusual RED triplet in its virion capsid. The quasispecies memory was eliminated when bottleneck events were so severe that they involved a number of particles close to or lower than the number of minority components of the mutant spectrum responsible for the molecular memory. This finding was documented by subjecting RED populations (that had reverted to the wild-type sequence) to passages at a very low multiplicity of infection. Also, individual viral clones from the population lost memory, as expected (Table 1). These results exclude the possibility that the frequent isolation of MAR mutants with the RED sequence (Table 1) was due to an inherent tendency of individual genomes harboring the RGD sequence in a particular context. Rather, the bottleneck experiments document that memory was a property of the population as a whole.
Equally complex are the evolutionary events involved in loss of the internal polyadenylate in clone C229. The initial clone included a highly heterogeneous polyadenylate tract (17), and fitness measurements indicated that the longer this polyadenylate tract, the lower the average fitness of the viral genome (18). The mutant spectrum of C229 p50, a population which underwent about 225 doublings in the amount of infectious particles from clone C229, included molecules with 1 and up to 10 additional adenylate residues (Table 2). In this case, memory involved retaining in the mutant spectrum of C229 p50 a repertoire of molecules that were themselves a modulated spectrum of that of the initial clone C229 (17).
It must be stressed that the isolation of FMDV RED or clones with an increased number of adenylate residues between genomic positions 1119 and 1123 could not be predicted by examination of the corresponding consensus sequences. Therefore, the frequent unpredictability of the types of genomes selected from different viral quasispecies subjected to identical constraints (antibodies, inhibitors, new susceptible host cell types, etc.) may be decisively influenced by the past evolutionary history of the viral quasispecies. In a number of natural infections with RNA viruses, minority components of quasispecies can reemerge, contributing to the maintenance of active viral replication in vivo (5, 6, 26, 36). A molecular memory provides the mechanisms for quasispecies evolving in vivo to respond efficiently to a selective pressure that has been previously experienced by the same population, even if this selective pressure had not been in operation for many viral generations. The influence of memory will be lost upon bottleneck transmissions but may be felt in the course of chronic infections or upon vertical or horizontal transmissions involving viral loads compatible with the carrying of minority components from the mutant spectra. Host-range mutants may be present as components of the quasispecies memory in an infected host. In this case, a population bottleneck during natural transmission of the virus may manifest the host-range variant by initiating an infection in an unusual host. In conclusion, we have documented the presence of a molecular memory in RNA viral quasispecies, a feature of biologically complex adaptive systems.
ACKNOWLEDGMENTS
We thank M. A. Rodríguez Marcos for valuable comments and M. Davila and G. Gómez-Mariano for expert technical assistance.
Our work was supported by grants from the DGES (PM97-0060-C02-01), FIS (98/0054-01), FAIR (5 PL97-3665), UE (PSS 0884), and Fundación Ramón Areces. C.M.R.-J. was supported by a predoctoral fellowship from CAM.
REFERENCES
- 1.Acharya R, Fry E, Stuart D, Fox G, Rowlands D, Brown F. The three dimensional structure of foot-and-mouth disease virus at 2.9 Å resolution. Nature. 1989;337:709–715. doi: 10.1038/337709a0. [DOI] [PubMed] [Google Scholar]
- 2.Ahmed R, Gray D. Immunological memory and protective immunity: understanding their relation. Science. 1996;272:54–79. doi: 10.1126/science.272.5258.54. [DOI] [PubMed] [Google Scholar]
- 3.Batschelet E, Domingo E, Weissmann C. The proportion of revertant and mutant phage in a growing population, as a function of mutation and growth rate. Gene. 1976;1:27–32. doi: 10.1016/0378-1119(76)90004-4. [DOI] [PubMed] [Google Scholar]
- 4.Berinstein A, Rovainen M, Hovi T, Mason R W, Baxt B. Antibodies to the vitronectin receptor (integrin αvβ3) inhibit binding and infection of foot-and-mouth disease virus. J Virol. 1995;69:2664–2666. doi: 10.1128/jvi.69.4.2664-2666.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borrow P, Lewicki H, Wei X P, Horwitz M S, Pfeffer N, Meyers H, Nelson J A, Gairin J E, Hahn B H, Oldstone M B A, Shaw G M. Antiviral pressure exerted by HIV-1 specific cytotoxic T lymphocytes (CTLs) during primary infection demonstrated by rapid selection of CTL escape virus. Nat Med. 1997;3:205–211. doi: 10.1038/nm0297-205. [DOI] [PubMed] [Google Scholar]
- 6.Briones C, Mas A, Gómez-Mariano G, Altisent C, Menéndez-Arias L, Soriano V, Domingo E. Dynamics of dominance of a dipeptide insertion in reverse transcriptase of HIV-1 from patients subjected to prolonged therapy. Virus Res. 2000;66:13–26. doi: 10.1016/s0168-1702(99)00120-3. [DOI] [PubMed] [Google Scholar]
- 7.Cline J, Braman J C, Hogrefe H H. PCR fidelity of pfu DNA polymerase and other thermostable DNA polymerases. Nucleic Acids Res. 1996;24:3546–3551. doi: 10.1093/nar/24.18.3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Domingo E, Holland J J. Mutation rates and rapid evolution of RNA viruses. In: Morse S S, editor. Evolutionary biology of viruses. New York, N.Y: Raven Press; 1994. pp. 161–184. [Google Scholar]
- 9.Domingo E, Escarmís C, Martínez M A, Martínez-Salas E, Mateu M G. Foot-and-mouth disease virus populations are quasispecies. Curr Top Microbiol Immunol. 1992;176:33–47. doi: 10.1007/978-3-642-77011-1_3. [DOI] [PubMed] [Google Scholar]
- 10.Domingo E, Sabo D L, Taniguchi T, Weissmann C. Nucleotide sequence heterogeneity of an RNA phage population. Cell. 1978;13:735–744. doi: 10.1016/0092-8674(78)90223-4. [DOI] [PubMed] [Google Scholar]
- 11.Domingo E, Webster R G, Holland J J, editors. Origin and evolution of viruses. San Diego, Calif: Academic Press; 1999. [Google Scholar]
- 12.Drake J W, Holland J J. Mutation rates among RNA viruses. Proc Natl Acad Sci USA. 1999;96:13910–13913. doi: 10.1073/pnas.96.24.13910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eigen M. Self-organization of matter and evolution of biological macromolecules. Naturwissenschaften. 1971;58:465–523. doi: 10.1007/BF00623322. [DOI] [PubMed] [Google Scholar]
- 14.Eigen M. Steps towards life. Oxford, United Kingdom: Oxford University Press; 1992. [Google Scholar]
- 15.Eigen M, Biebricher C K. Sequence space and quasispecies distribution. In: Domingo E, Holland J J, Ahlquist P, editors. RNA genetics. Vol. 3. Boca Raton, Fla: CRC Press, Inc.; 1988. pp. 211–245. [Google Scholar]
- 16.Eigen M, Schuster P. The hypercycle. A principle of natural self-organization. Berlin, Germany: Springer-Verlang; 1979. [DOI] [PubMed] [Google Scholar]
- 17.Escarmís C, Dávila M, Charpentier N, Bracho A, Moya A, Domingo E. Genetic lesions associated with Muller's ratchet in an RNA virus. J Mol Biol. 1996;264:255–267. doi: 10.1006/jmbi.1996.0639. [DOI] [PubMed] [Google Scholar]
- 18.Escarmís C, Dávila M, Domingo E. Multiple molecular pathways for fitness recovery of an RNA virus debilitated by operation of Muller's ratchet. J Mol Biol. 1999;285:495–505. doi: 10.1006/jmbi.1998.2366. [DOI] [PubMed] [Google Scholar]
- 19.Frank S A. The design of natural and artificial adaptive systems. In: Rose M R, Lauder G V, editors. Adaptation. San Diego, Calif: Academic Press; 1996. pp. 451–505. [Google Scholar]
- 20.Gell-Mann M. Complex adaptive systems. In: Cowan G A, Pines D, Meltzer D, editors. Complexity. Metaphors, models, and reality. Reading, Mass: Addison-Wesley Publishing Co.; 1994. pp. 17–45. [Google Scholar]
- 21.Holland J J, de la Torre J C, Clarke D K, Duarte E. Quantitation of relative fitness and great adaptability of clonal populations of RNA viruses. J Virol. 1991;65:2960–2967. doi: 10.1128/jvi.65.6.2960-2967.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holland J J, de la Torre J C, Steinhauer D. RNA virus populations as quasispecies. Curr Top Microbiol Immunol. 1992;176:1–20. doi: 10.1007/978-3-642-77011-1_1. [DOI] [PubMed] [Google Scholar]
- 23.Jackson T, Sharma A, Abu Ghazaleh R, Blakemore W E, Ellard F M, Simmons D F L, Newman J W I, Stuart D I, King A M Q. Arginine-glycine-aspartic acid-specific binding by foot-and-mouth disease virus to the purified αvβ3 integrin in vitro. J Virol. 1997;71:8357–8361. doi: 10.1128/jvi.71.11.8357-8361.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jacob J, Baltimore D. Modelling T-cell memory by genetic marking of memory T cells in vivo. Nature. 1999;399:593–597. doi: 10.1038/21208. [DOI] [PubMed] [Google Scholar]
- 25.Jerne N K. The natural selection theory of antibody formation. Proc Natl Acad Sci USA. 1955;41:849–857. doi: 10.1073/pnas.41.11.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karlsson A C, Gaines H, Sällberg M, Lindbäck S, Sönnerborg A. Reappearance of founder virus sequence in human immunodeficiency virus type 1-infected patients. J Virol. 1999;73:6191–6196. doi: 10.1128/jvi.73.7.6191-6196.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martínez M A, Verdaguer N, Mateu M G, Domingo E. Evolution subverting essentiality: dispensability of the cell attachment Arg-Gly-Asp motif in multiply passaged foot-and-mouth disease virus. Proc Natl Acad Sci USA. 1997;94:6798–6802. doi: 10.1073/pnas.94.13.6798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mateu M G, Martínez M A, Rocha E, Andreu D, Parejo J, Giralt E, Sobrino F, Domingo E. Implications of a quasispecies genome structure: effect of frequent, naturally occurring amino acid substitutions on the antigenicity of foot-and-mouth disease virus. Proc Natl Acad Sci USA. 1989;86:5883–5887. doi: 10.1073/pnas.86.15.5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murphy F A. Virus taxonomy. In: Fields B N, et al., editors. Fields virology. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 15–57. [Google Scholar]
- 30.Rueckert R R. Picornaviridae: the viruses and their replication. In: Fields B N, et al., editors. Fields virology. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 609–654. [Google Scholar]
- 31.Ruiz-Jarabo C M, Sevilla N, Dávila M, Gómez-Mariano G, Baranowski E, Domingo E. Antigenic properties and population stability of a foot-and-mouth disease virus with an altered Arg-Gly-Asp receptor-recognition motif. J Gen Virol. 1999;80:1899–1909. doi: 10.1099/0022-1317-80-8-1899. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 33.Sobrino F, Dávila M, Ortín J, Domingo E. Multiple genetic variants arise in the course of replication of foot-and-mouth disease virus in cell culture. Virology. 1983;128:310–318. doi: 10.1016/0042-6822(83)90258-1. [DOI] [PubMed] [Google Scholar]
- 34.Verdaguer N, Mateu M G, Andreu D, Giralt E, Domingo E, Fita I. Structure of the major antigenic loop of foot-and-mouth disease virus complexed with a neutralizing antibody: direct involvement of the Arg-Gly-Asp motif in the interaction. EMBO J. 1995;14:1690–1696. doi: 10.1002/j.1460-2075.1995.tb07158.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Verdaguer N, Sevilla N, Valero M L, Stuart D, Brocchi E, Andreu D, Giralt E, Domingo E, Mateu M G, Fita I. A similar pattern of interaction for different antibodies with a major antigenic site of foot-and-mouth disease virus: implications for intratypic antigenic variation. J Virol. 1998;72:739–748. doi: 10.1128/jvi.72.1.739-748.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wyatt C A, Andrus L, Brotman B, Huang F, Lee D-H, Prince A M. Immunity in chimpanzees chronically infected with hepatitis C virus: role of minor quasispecies in reinfection. J Virol. 1998;72:1725–1730. doi: 10.1128/jvi.72.3.1725-1730.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]