Figure 1. Reference mapping model accurately predicts malignant CD4+ T cells in classic MF.
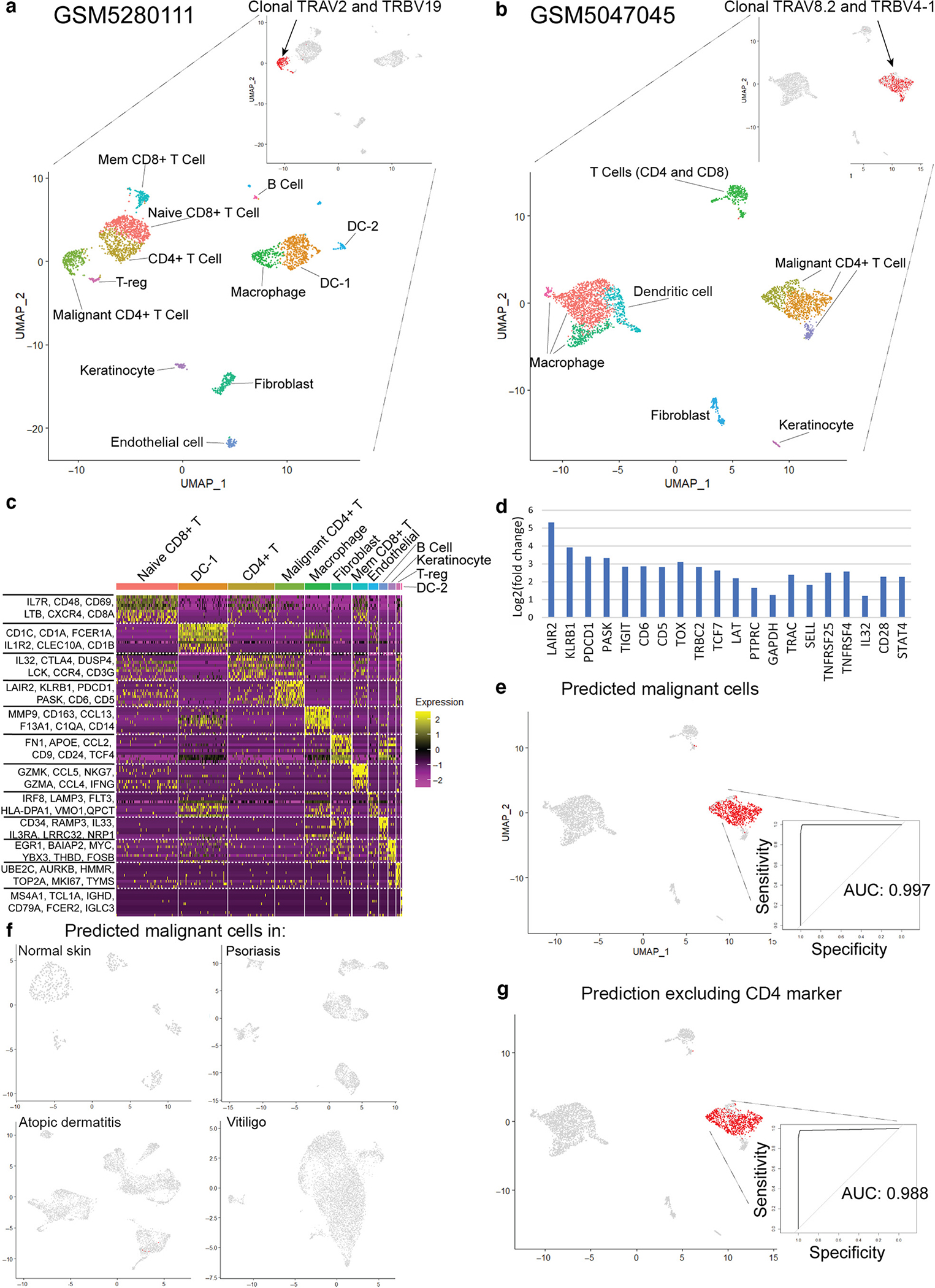
(a) UMAP of targeted scRNA-seq in one patient with classic MF (GEO accession number GSM5280111) and identification of malignant and nonmalignant cell types of the tumor microenvironment. (b) UMAP of scRNA-seq in a second patient with classic MF (GEO accession number GSM5047045). (c) Heatmap of cell clusters in GSM5280111. Genes with the greatest differential expression in each region are listed. (d) Identification of the top 20 differentially expressed genes of the malignant CD4+ T-cell population in GSM5280111. (e) Reference mapping prediction of malignant cells in GSM5047045 using a model built on GSM5280111. (f) Reference mapping prediction of malignant cells in normal skin, psoriasis, atopic dermatitis, and vitiligo samples using the model. (g) Reference mapping prediction of malignant cells in GSM5047045, excluding CD4 from the model. AUC, area under the curve; DC, dendritic cell; GEO, Gene Expression Omnibus; MF, mycosis fungoides; scRNA-seq, single-cell RNA sequencing; T-reg, regulatory T cell; UMAP, Uniform Manifold Approximation and Projection.
