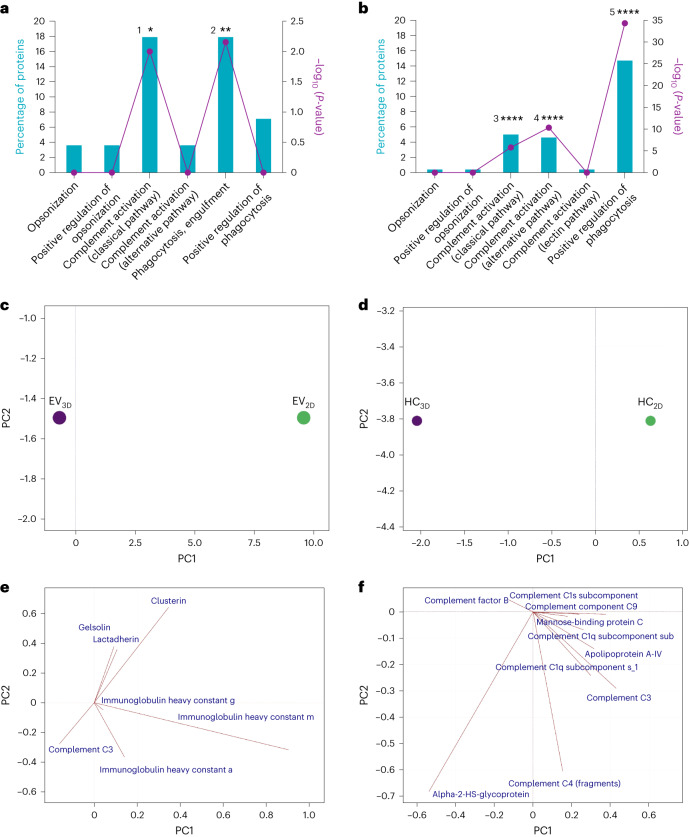
Fig. 5. GO analysis for classification of the protein identified by LC–MS.
a,b, Proteins detected in both EV3D/2D and HC2D/3D were decoded to obtain a gene list involved in biological processes contributing to the clearance of non-incubated EVs (against in-built UniProt human database, GO analysis with Bonferroni correction (1*P = 0.01 and 2**P = 0.007) (a) and hard protein corona of incubated EVs (against in-built UniProt non-human mammal (bovine) database, GO analysis with Bonferroni correction, 3****P = 1.6 × 10−6, 4****P = 4.97 × 10−11 and 5****P = 4.89 × 10−35) (b). c,d, GO analysis was performed using FunRich software v.3.1.3. Significantly enriched proteins (hypergeometric and Bonferroni analysis *P < 0.05, **P < 0.01 and ****P < 0.0001) by quantity underwent PCA for separation of EV2D and EV3D (c) and HC2D and HC3D (d), shown as score plots. e,f, Loading plots were used to illustrate how each protein influences the computed PCs in c,d, respectively. Overall, the intrinsic properties of EV protein corona constituents are fundamentally different. HC2D proteins are associated with higher extents of complement activation. The PCA was performed using mean of the LC–MS data (two biological and three technical replicates), and EVs were pooled from three batches.