Fig. 1. Identification of base-editable CD45 variants with stable biophysical properties.
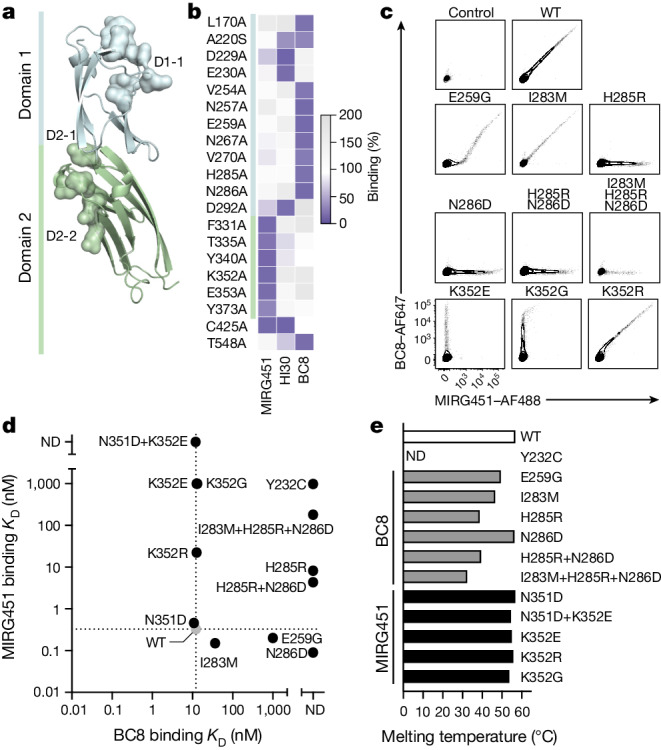
a, Crystal structure (retrieved from PDB under accession number 5FMV) of the CD45 domain 1 (blue) and domain 2 (green) highlighting selected regions D1-1, D2-1 and D2-2 as surface density. b, Heatmap showing CD45 alanine substitutions that result in less than 20% residual binding for one monoclonal antibody but have more than 70% for either of the other two tested anti-CD45 antibodies. c, Flow cytometry of CD45 variants overexpressed in DF-1 cells affecting the MIRG451 and BC8 epitopes (n = 4 biological replicates). WT, wild type. d, MIRG451 and BC8 binding affinity for purified recombinant CD45 D1–D2 protein variants measured by BLI. The grey circle refers to the wild type. KD, dissociation constant; ND, not detected. e, Melting temperature of purified recombinant CD45 D1–D2 protein variants (n = 1–2).
