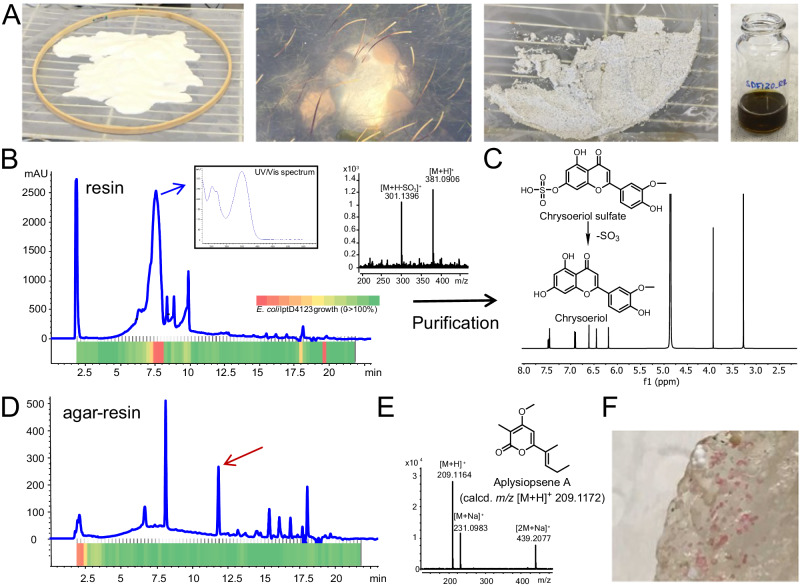
Fig. 1. SMIRC deployment in a Zostera marina meadow (Mission Bay, San Diego), extract analysis, compound identification, and in situ microbial growth.
A From left to right: activated HP-20 resin before enclosing in Nitex mesh and embroidery hoop, SMIRC deployment, recovered resin before extraction, concentrated extract in MeOH. B UV360 chromatogram of the resin extract and corresponding microfractionation bioassay (80 fractions collected over 20 min) with heatmap showing E. coli LptD4123 growth inhibition (OD650, red = no growth, green = 100% growth relative to control). Blue arrow: UV spectrum of active peak (7.6 min). MS spectrum of the active compound shows loss of sulfate. C 1H NMR (500 MHz) and structure of the flavonoid chrysoeriol, a degradation product of chrysoeriol sulfate. D UV360 chromatogram of extract generated from resin embedded in agar (in situ cultivation) deployed at the same site revealed an additional peak (red arrow). E Structure and MS spectrum of aplysiopsene A isolated from the agar/resin peak (red arrow). F Pink colonies growing on agar/resin matrix. UV (ultraviolet), calcd (calculated), OD (optical density).