Figure 2.
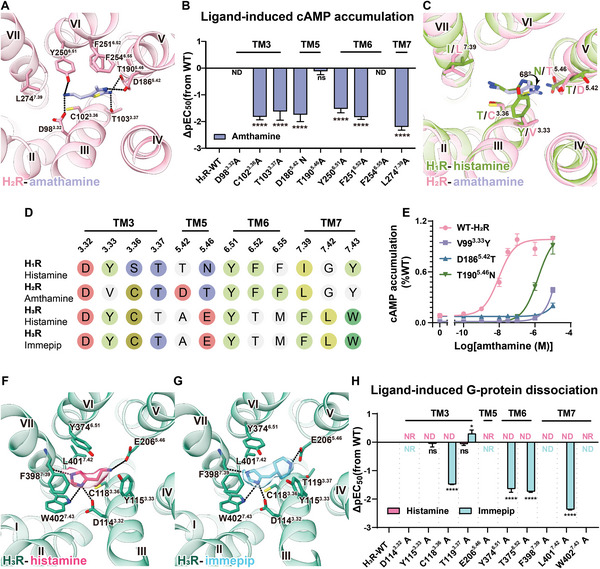
Ligand recognition of histamine receptors. A) Detailed interaction of amthamine with H2R. Dashed lines represent hydrogen bonds. B) Amthamine induced cAMP accumulation in HEK293 cells expressing H2R mutants of the residues in orthosteric pocket (n = 3, ordinary one‐way ANOVA, ****P < 0.0001, ND, not determinable, which refers to cannot be established over the tested concentration range, ns refers to no significance between the WT and mutant). C) Structural comparisons of ligand recognition between histamine‐H1R and amthamine‐H2R structures. D) Sequence alignment of the orthosteric pocket of histamine receptors. Residues that interact with ligands are highlighted with colored circles. E) Dose‐dependent curves for amthamine induced cAMP accumulation in HEK293 cells expressing the H2R mutants (V993.33Y, D1865.42T, T1905.46N) that are not conserved in H1R (n = 3). F‐G) Detailed interactions of histamine F) and immepip G) with H3R. H) Agonists induced Gi dissociation in HEK293 cells expressing H3R mutants of the residues in orthosteric pocket by NanoBiT assays (n = 3, ordinary one‐way ANOVA, *P = 0.0456, ****P < 0.0001, NR refers to no response to the ligand, ND, not determinable, which refers to cannot be established over the tested concentration range, ns refers to no significance between the WT and mutant).
