Figure 1: Architecture and example code for SOURSOP.
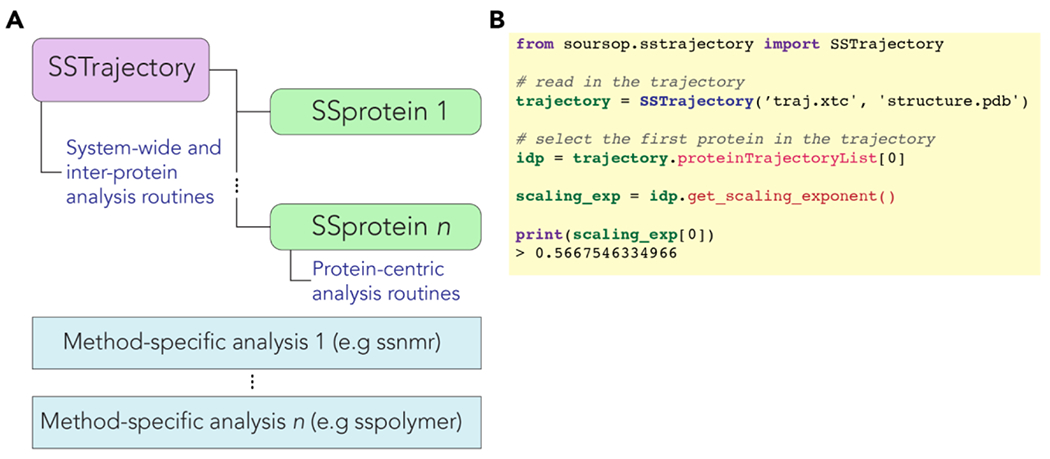
(A) Trajectory files are read into an SSTrajectory object. This object automatically parses each polypeptide chain into separate SSProtein objects. Each SSProtein object has a set of object-based analyses associated with them. Each trajectory must have between 1 and protein chains in it. In addition, various stateless method-specific analysis modules exist for certain types of analysis. Additional stateless methods can be extended to allow new analysis routines to be incorporated in a way that does not alter the SSProtein or SSTrajectory code. (B) Example code illustrating how the apparent scaling exponent can be calculated from an ensemble.
