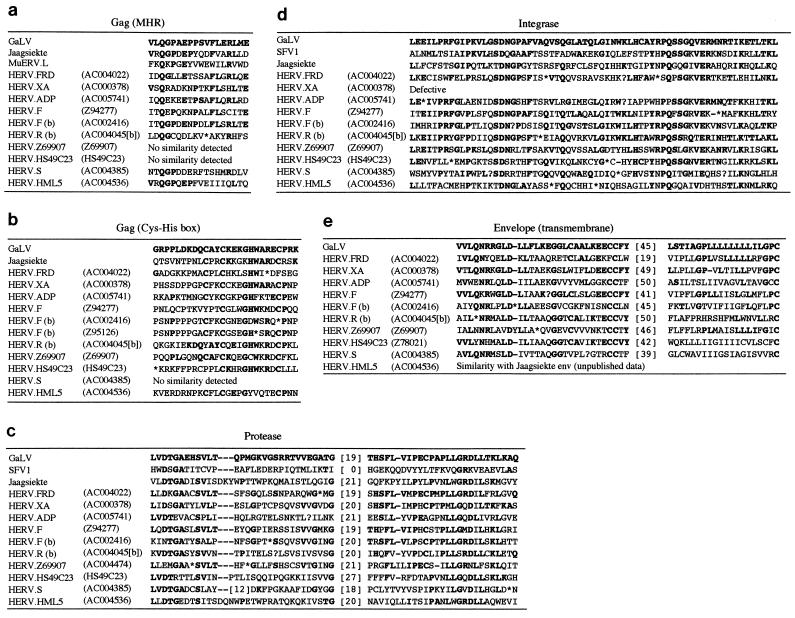
FIG. 4.
Alignment of conserved amino acid motifs present within the HERV families identified in this report. The figures in parentheses indicate the cosmids from which the sequences were derived. GaLV, Jaagsiekte, HSV, and MuERV.L are provided for comparative purposes. (a) Gag alignment spanning nucleotide positions 1660 to 1713 within the GaLV sequence (13); (b) Gag alignment spanning positions 2074 to 2157; (c) Pro alignment spanning positions 2266 to 2490; (d) Int alignment spanning positions 4969 to 5166; (e) Env alignment spanning positions 7220 to 7504.