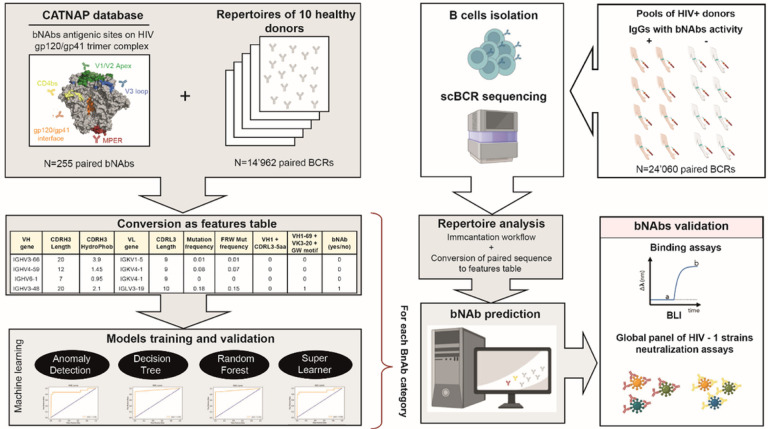
Figure 1. RAIN pipeline for automatic identification of bNAbs.
Data collected from the CATNAP database (bNAbs) and healthy donor repertoires (mAbs) were converted as a feature table to train and validate four machine learning models: anomaly detection (AD), decision tree (DT), random forest (RF) and super learner (SL). We performed single-cell BCR sequencing from HIV-1 seropositive donors with (illustrated by orange arm) or without (illustrated by white arm) sera broadly neutralizing activities. BCR sequences are processed by Immcantation workflow and analyzed as a features table. Next, the predicted bNAbs found by the four algorithms were produced and tested in neutralization and binding assays.