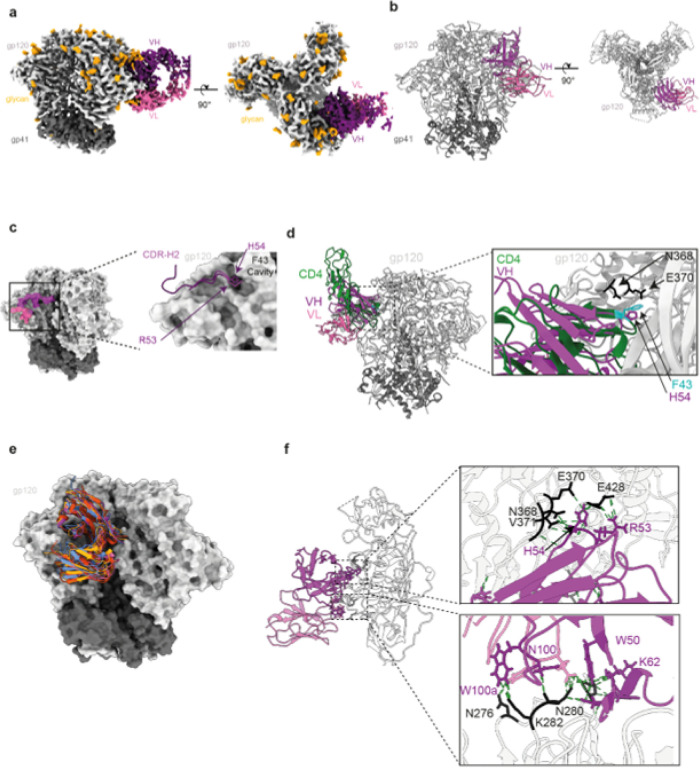
Figure 5. Cryo-EM map and structure of Fab SOSIP complexes.
(a)-Side and top views of the cryo-EM density map of the Fab4251-DS-SOSIP complex, with gp120 in light grey, gp41 in dark gray, VH in violet and VL in pink. (b)-Atomic model of Fab4251-DS-SOSIP complex shown in cartoon representation. (c)-Foot print representation of the heavy and light chain binding surface on DS-SOSIP, colored according to a. Inlet on the right in represent the HCDR2 loop in violet, with H54 in the Phe-43 cavity. (d)-Overlay of CD4 receptor (green) bound to SOSIP (PDB.5U1F) and Fab4251 (violet). Inlet highlights positions N368, E370 on gp120 and F43 on CD4 and H54 of the VH. (e)-Overlay of VRC01 class antibodies on SOSIP with Fab4251 (violet), VRC01 (PDB.6V8X, green), PG04 (PDB.4I3S, red), and 3BNC60 (PDB.4GW4, orange). (f)-Contact residues at the Fab4251-gp120 interface. Contact residues are defined as two residues containing any atom within 4 Å of each other.