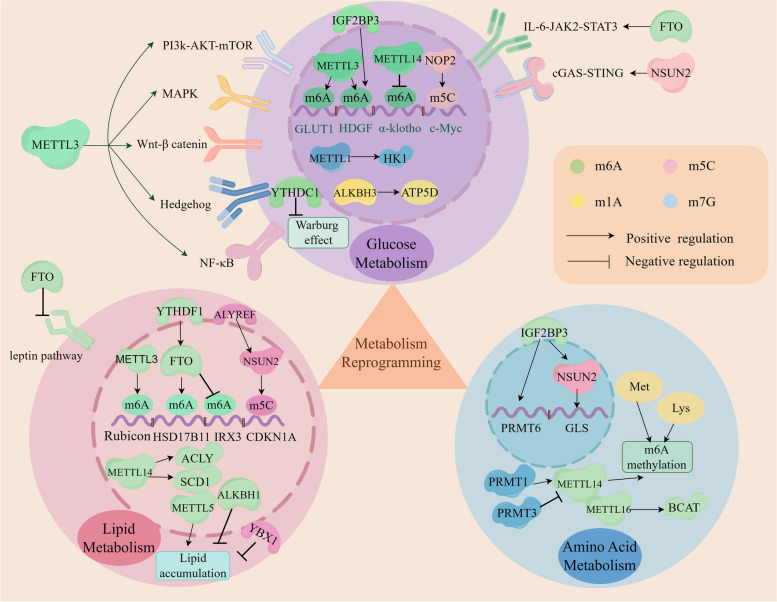
Fig. 3.
RNA methylations participate in metabolic reprogramming of the TME, including glucose metabolism, lipid metabolism and amino acid metabolism. RNA methylations regulate the expression of glycolysis-associated genes (GLUT1, Gys2, HDGF) and signal pathways (PI3K-AKT, mTORC1, MAPK, Wnt-β catenin, Hedgehog, NF-κB, IL-6/JAK2/STAT3, cGAS/STING) and enhance Warburg effect through their regulators, such as METTL1, METTL3, METTL14, NOP2, NSUN2, FTO, ALKBH3, IGF2BP3, YTHDC1 and. m6A and m5C accelerate lipid accumulation. m6A, m5C and m7G modulate the metabolisms of glutamine, arginine, methionine and lysine. These methylations impact tumor cell immunogenicity, proliferation, immune escape as well as tumor progression. ACLY, ATP citrate lyase; SCD1, stearoyl-CoA desaturase1; BCAT1, branched-chain amino acid transaminase 1; Met, methionine; Lys, lysine; PRMT1, protein arginine methyltransferase1. Figure created with figdraw.com