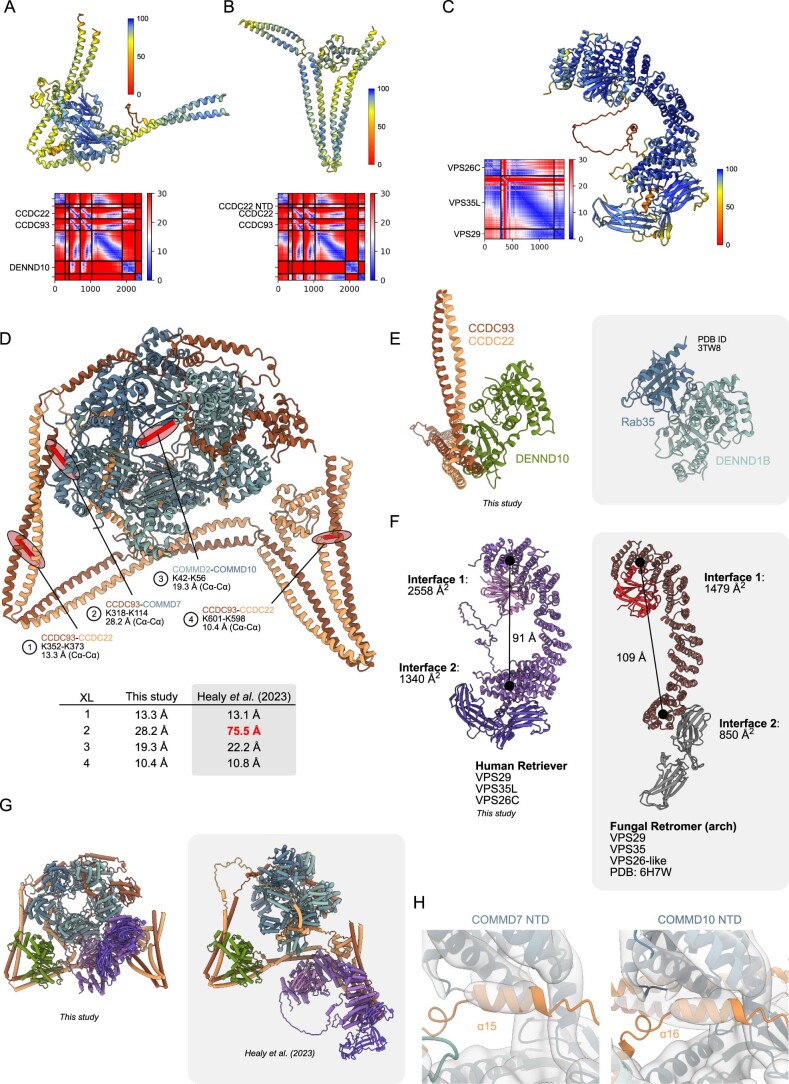
Extended Data Fig. 4. Molecular models of Commander complex bottom half, related to Fig. 4.
a, AF2 model of DENND10 region of the Commander complex used as an initial model with predicted alignment error plots indicating the relevant chains. Model is colored according to per-residue pLDDt scores. Model has been trimmed based on the fit to the cryo-EM density map. AF2 model contained all chains of the bottom half during prediction. b, V-coil region of Commander as in a, with different random seed in AF2 prediction. c, Retriever subcomplex model as in a. d, Chemical crosslinks identified by MS in the context of the Commander structure model. e, Comparison of DENND1B-Rab35 complex structure (PDB 3TW8) with DENND10 in the context of Commander. I-coil sterically blocks the putative Rab binding site on DENND10. f, Structure of Retriever in the context of the Commander complex compared to the Fungal retromer structure. Interface 1: VPS29-VPS35(L). Interface 2: VPS35(L)-VPS26(C). Retriever adopts a contracted conformation compared to retromer and exhibits larger interaction interfaces. g, Comparison of Commander complex models presented in this study (left) and in Healy et al. (right), kindly provided by the authors, superposed via DENND10. h, Density supporting the placement of CCDC22 α15 and α16. Map (EMD-17340) was low-pass filtered to 7 Å.