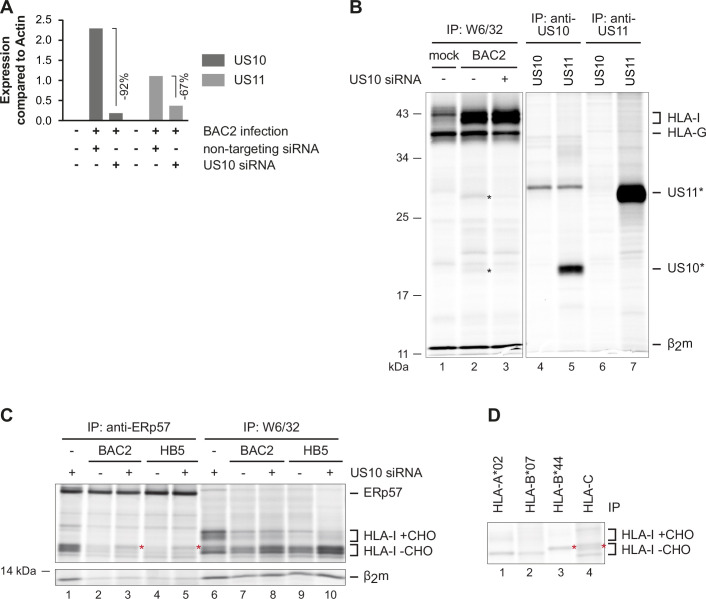
Figure 6. Downregulation of overlapping US10 and US11 transcripts in human cytomegalovirus (HCMV)-infected cells rescues human leucocyte antigen class I (HLA-I) interaction with the peptide loading complex (PLC).
(A) MRC-5 fibroblasts were nucleofected with US10-specific or non-targeting siRNA 24 hr prior to mock treatment or infection with HCMV ΔUS2-6 mutant BAC2 at an MOI (multiplicity of infection) of 5. At 24 hr p.i., RNA was isolated. Subsequently, cDNA was generated and analyzed by qPCR. The binding sites for the used primers are depicted in Figure 6—figure supplement 2. Expression of US10 and US11 is shown compared to expression of actin. (B) HLA-G-expressing BJ-5ta fibroblasts were treated with siRNA and infected as in (A). At 48 hr post-infection, cells were metabolically labeled for 2 hr. Digitonin cell lysates were prepared, and immunoprecipitations were performed as indicated (lanes 1–3). In parallel, immunoprecipitations were performed with HeLa cells transfected with US10 or US11 expression plasmids, lanes 4–7 (Figure 6—source data 1). (C) MRC-5 fibroblasts were nucleofected with US10-specific or non-targeting siRNA 24 hr prior to mock treatment or infection with the HCMV ΔUS2-6 mutants BAC2 or HB5 at an MOI of 7. Proteins were metabolically labeled at 24 h p.i. for 2 hr, and immunoprecipitations using anti-ERp57 or W6/32 were performed. All samples were treated by EndoH (Figure 6—source data 2). Asterisk: strongly increased HLA-I HC when applying US10 siRNA. One of two independent experiments (biological replicates) is shown. (D) BAC2-infected MRC-5 fibroblasts were treated as in (C), and HLA-I-specific immunoprecipitations were performed as indicated. All samples were EndoH-treated prior to separation by SDS-PAGE (Figure 6—source data 2).