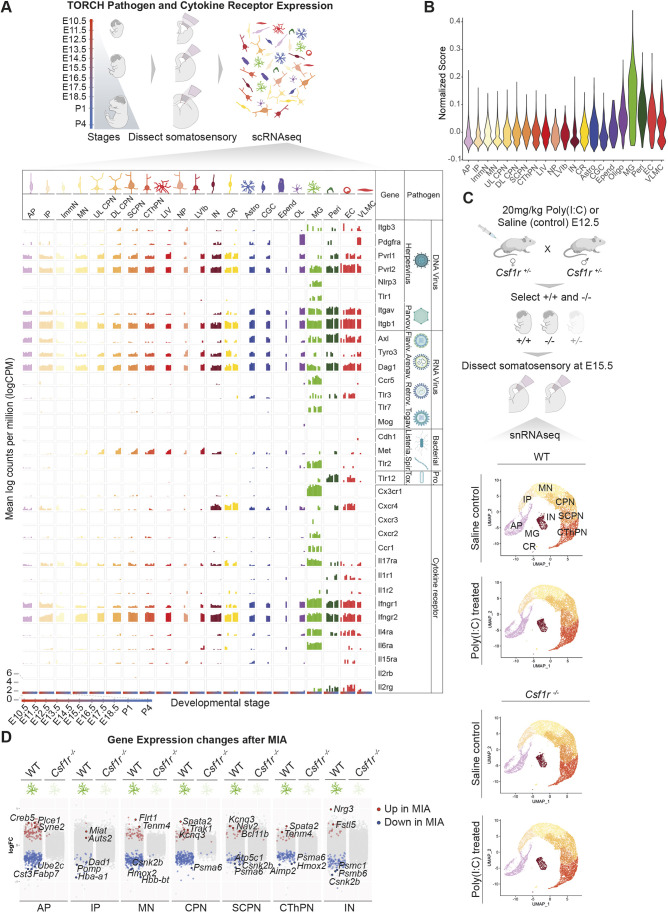
Fig. 1.
Brain transcriptional changes after MIA require microglia. (A) Simplified schematic of sample processing pipeline from Di Bella et al. (2021) and bar plots of TORCH pathogen and cytokine receptor expression (mean log counts per million) in each cell type across development. (B) Violin plot of CytoTORCH module score for each cell cluster (MG, P<0.05 for ANOVA and for pairwise comparisons between microglia and all other cell types using Tukey's HST). (C) Overview of experimental design (top) and UMAP visualization of snRNAseq data from the developing somatosensory cortex of E15.5 wild-type and Csf1r−/− mice after saline or Poly(I:C) maternal injection at E12.5 (two or three embryos per condition from a total of four pregnant dams, 42,736 total cells) (bottom). Cells are colored by cell type assignment. (D) Dot plots of log(fold change) in gene expression between MIA and saline for cell types with significant changes at E15.5. DEG analysis was performed using NEBULA. AP, apical progenitors; IP, intermediate progenitors; ImmN, immature neurons, MN, migrating neurons; (UL/DL) CPN, (upper layer/deep layer) callosal projection neurons; SCPN, subcerebral projection neurons; CThPN, corticothalamic projection neurons; LIV, layer IV stellate neurons; IN, interneurons; CR, Cajal-Retzius cells; Astro, astrocytes; CGC, cycling glial cells; Epend; ependymal cells; OL, oligodendroglia; MG, microglia; Peri, pericytes; EC, endothelial cells; VLMC, vascular and leptomeningeal cells. Created with BioRender.com.