Fig. 7.
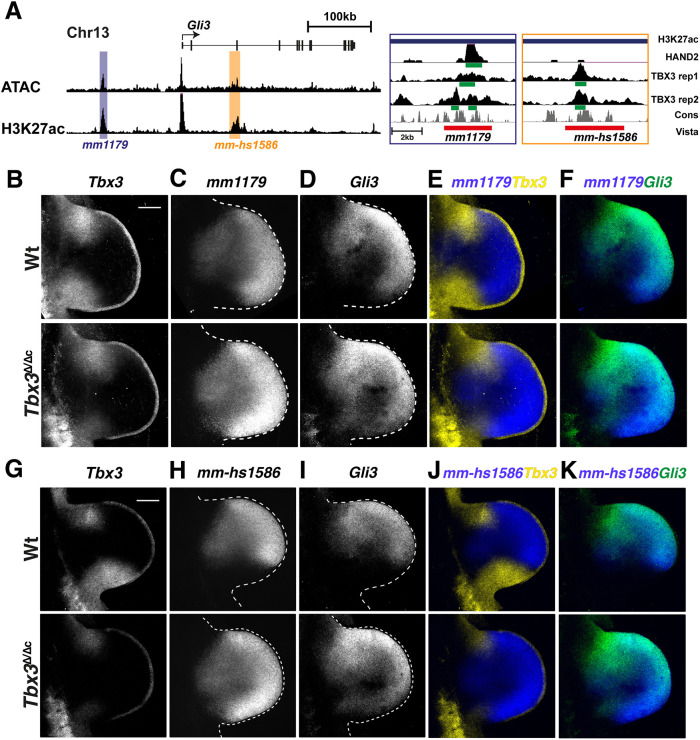
Tbx3 is required to restrict the activity of the Gli3 limb enhancers and Gli3 expression from the posterior limb bud mesenchyme. (A) UCSC browser view of parts of the Gli3 genomic landscape with regions of accessible and active chromatin tracks (ATAC-seq and H3K27ac peaks; Malkmus et al., 2021). The two limb enhancers mm1179 and mm-hs1586 are highlighted. The enlargements on the right show the genomic regions tested for Gli3 enhancer activity (red bar) and the HAND2 and TBX3 ChIP-seq peaks. The green bars indicate the called peaks. Both replicates for the TBX3 ChIP-seq are shown; the HAND2 ChIP-seq data is from Osterwalder et al. (2014). (B-K) RNA-FISH analysis of wild-type (Wt) and Tbx3Δ/Δc mouse limb buds (E10.5, 35-37 somites) of endogenous Tbx3 (yellow in E,J), transgenic enhancer-lacZ reporter RNA expression (blue in E,F,J,K; mm1179 in C,E,F and mm-hs1586 in H,J,K) and endogenous Gli3 expression (green in F,K). n=5 biological replicates were analysed per probe and genotype. Note: low and variable levels of remaining non-functional Tbx3 Δ transcripts are detected in Tbx3Δ/Δc limb buds by the HCRTM probe set. All forelimb buds are oriented with anterior to the top and posterior to the bottom. Dashed lines delineate the shape of the limb bud. Scale bars: 200 µm.