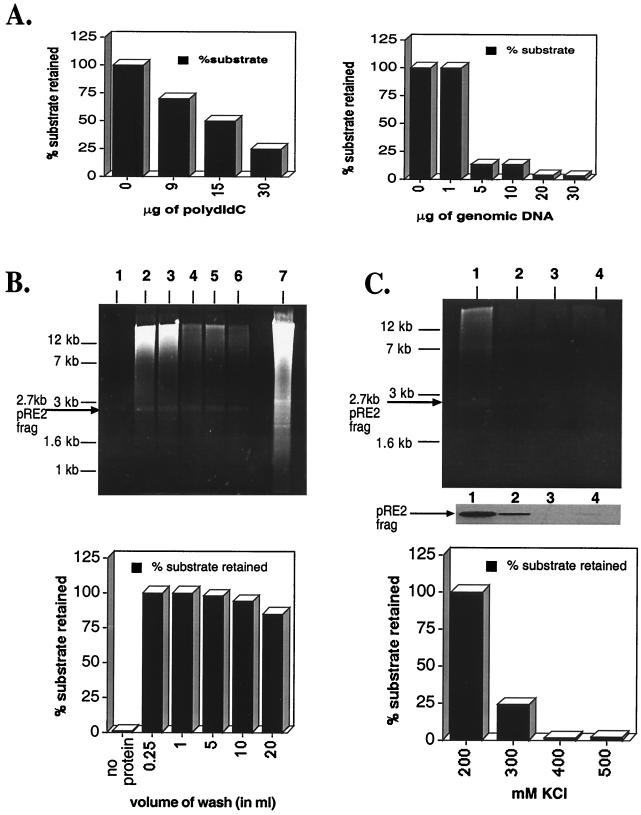
FIG. 2.
Optimization of filter-binding reactions. (A) The left panel is a titration of increasing amounts of poly(dI-dC) required to disrupt Rep–ch-19 RBE-specific binding (as measured by filter binding). The reaction contains 20 pmol of Rep68H6 and 1 fmol of the labeled ch-19 target sequences. The right panel graphs the effect of increasing amounts of genomic DNA on Rep68H6–ch-19-labeled DNA as measured by the filter binding assay (see Materials and Methods for details). (B) Titration of nonelution buffer on Rep–ch-19-specific binding by ethidium bromide staining of agarose gel analysis (upper panel) and retention of radio labeled ch-19-specific probe (lower panel). Lane 1, no Rep68H6 protein; lanes 2 to 6, 0.25, 1.0, 5.0, 10, and 20 ml of nonelution wash buffer, respectively. Lane 7 shows 10 μg of BamHI-digested HeLa genomic DNA prior to filter binding and elution. In all the reactions, 50 fmol of the 32P-labeled ch-19 fragment from pRE2 was spiked in to serve as an internal control of Rep-specific binding. Results are graphed in the lower panel. (C) KCl effect on Rep genomic DNA filter-binding assay using ethidium bromide staining of agarose gel analysis (upper panel) and retention of radio labeled ch-19-specific probe (lower panel). Lane 1, 200 mM KCl; lane 2, 300 mM KCl; lane 3, 400 mM KCl; lane 4, 500 mM KCl. Reactions were carried out as described in panel A and in Materials and Methods and was followed by washing with 10 ml of nonelution buffer. The middle part of panel C shows an autoradiogram of retained ch-19 fragment. The lower panel shows a graph of the percentage of the ch-19 fragment retained with respect to increasing amounts of KCl.