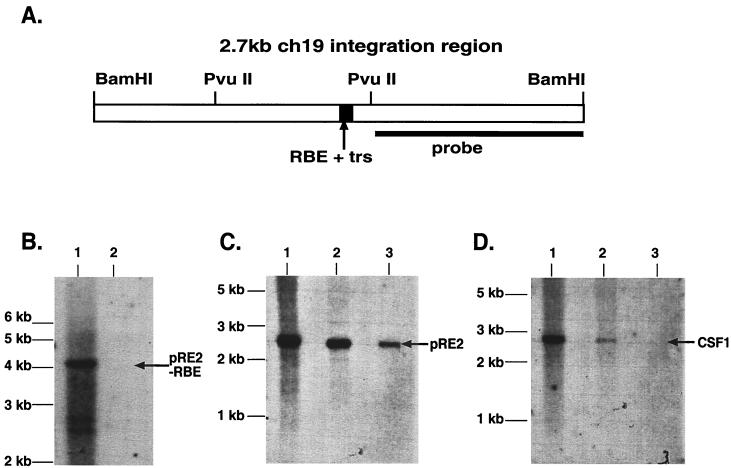
FIG. 3.
Southern Blot analysis of filter-bound genomic DNA. (A) Diagram of the 2.7-kb ch-19 integration region. The RBE-trs is labeled and shown as a black box, while the sequence recognition region of the probe used for all analysis is depicted as a solid line. The distance between the two PvuII sites is 898 bp. (B) Filter-binding assay of ch-19 sequences devoid of any known RBE sequences. Genomic DNA digested with PvuII (one site in the 2.7-kb fragment and the other 3′ to the BamHI site) is specific for a 4-kb ch-19 fragment devoid of RBE sequences. Lane 1, signal generated with 10 μg of PvuII-digested HeLa genomic DNA prior to filter binding assay; lane 2, signal generated after loading DNA eluted from one filter-binding reaction. Hybridization was carried out with the PvuII-BamHI right-side-specific probe (see panel A). (C) Southern analysis demonstrating Rep68H6's ability to retain ch-19 sequences carrying RBE sequences after the filter-binding assay. Lane 1, signal generated from 10 μg of BamHI-digested HeLa genomic DNA prior to filter binding; lane 2, signal generated from four pooled filter-binding reactions; lane 3, signal generated from one filter-binding reaction. The arrow indicates the position of the ch-19 fragment (pRE2). (D) Rep68H6 retention of non-ch-19 RBE genomic sequences. Lane 1, signal generated from 10 μg of BamHI-digested HeLa genomic DNA prior to filter-binding reaction; lane 2, signal generated from four pooled filter-binding reactions; lane 3, signal generated from one filter-binding reaction. The blot was probed with a SmaI-PstI fragment from pcCSF17, which recognizes exon 1 from the CSF1 gene (see Materials and Methods for details). The arrow indicates the position of the CSF1 fragment.