Abstract

Microtubule affinity-regulating kinase 4 (MARK4) is a serine-threonine kinase that phosphorylates microtubule-associated proteins (MAPs) and increases the microtubule dynamics. Due to its direct involvement in initiation, cell division, progression, and cancer metastasis, MARK4 is considered a potential therapeutic target. Here, we designed, synthesized, and characterized vanillin-isatin hybrids and evaluated their MARK4 inhibitory potential. All of the compounds strongly bind to MARK4 and interact closely with the active site residues. Finally, the compound VI-9 was selected for further investigation due to its high binding affinity and strong MARK4 inhibitory potential. Tau-phosphorylation assay has further confirmed that VI-9 significantly reduced the activity of MARK4. Compared with vanillin, VI-9 showed a better binding affinity and MARK4 inhibitory potential. Cell viability assays on human hepatocellular carcinoma (HCC) cell lines C3A and SNU-475 revealed that VI-9 inhibited their growth and proliferation. In addition, these compounds were nontoxic (up to 200 μM) for noncancerous (HEK-293) cells. Interestingly, VI-9 induces apoptosis and decreases the metastatic potential of the C3A and SNU-475 cell lines. The present work opens a newer avenue for vanillin-isatin hybrids and their derivatives in developing MARK4-targeted anticancer therapies.
1. Introduction
Microtubule affinity-regulating kinase 4 (MARK4) is a serine-threonine kinase and a promising drug target for diabetes, cancer, and neurological disorders due to its direct role in controlling microtubule dynamics. It phosphorylates microtubule-associated proteins (MAPs), such as tau, causing these proteins to separate from microtubules and enhancing microtubule dynamics.1,2 MARK4 is overexpressed in glioma cells, neural progenitor cells, hepatocellular carcinoma (HCC), and breast and lung cancers, suggesting its possible involvement in the initiation and advancement of cancer.3 Magnani et al.4 show the role of MARK4 in the modulation of glioblastoma stem cells as it is involved in the differentiation and proliferation of neural stem cells.4 MARK4 is specifically localized in the centrosome and microtubules.2 In normal glioma cells, MARK4 is located near the midbody and associated with centrosomes.4,5 MARK4 has its role in the cytoskeletal dynamics and cell division of malignant and nonmalignant cells. HCC, glioma, metastatic breast cancer, Alzheimer’s disease, and various metabolic disorders are associated with the aberrant expression of MARK46−8 This makes MARK4 a promising therapeutic target.9,10
The development of novel and promising therapeutic molecules involves the integration of multiple pharmacophoric subunits into a single scaffold using the molecular hybridization technique, as it was predicted that combining multiple physiologically active components would result in a potential chemotherapeutic entity. isatin (1H-indole-2,3-dione), a heterocyclic scaffold that Erdman and Laurent first isolated in 1840, is a pharmacophore with enormous biological activities.11 It is a metabolic byproduct of adrenaline in blood, the central nervous system and body fluids.12 Isatin derivatives showed MARK4 inhibitory effects, making them potential candidates for developing anticancer drugs.13 In preclinical studies, isatin and its derivatives demonstrated promising antiproliferative and antimetastatic effects, indicating their potential utility in cancer therapy.13 Thus, further research and clinical trials are necessary to fully elucidate its efficacy and safety profile in cancer treatment fully.
Isatin is an important scaffold present in the parotid gland secretions of Bufo frogs. Isatin is also a component of the FDA-approved anticancer drug [Sunitinib (SUN/SU11248)] used to treat renal cell carcinoma.14 Even though a variety of isatin-derived compounds that inhibit tyrosine and serine/threonine kinases are available, including SU4984 and SU5416, the clinical development of nintedanib (BIBF1120)15 and their molecular structures are drawn from ChemDraw, details of the compound were taken from Pubchem (Figure 1). An additional isatin-based triple angiokinase inhibitor is presently being studied in phase III clinical trials to treat nonsmall cell lung cancer (NSCLC).16
Figure 1.
Structure of a few FDA-approved anticancer drugs containing an isatin core.
HCC is one of the leading causes of cancer-related deaths worldwide.17 HCC may arise because of hepatitis B or C, cirrhosis, and other liver diseases induced by aflatoxin toxicity and metabolic dysregulation.18,19 The process of hepatocarcinogenesis involves an initial genotoxic insult (initiation), clonal expansion from precancerous to cancerous lesions (promotion), and an ultimate tumor spread through clonal expansion.20,21 The observed overexpression of MARK4 in multiple cancers, including HCC, justifies its characterization as a potential target for cancer treatment. Previously, we have shown that isatin-triazole hydrazones and vanillin-triazoles potentially inhibited MARK4 activity, leading to cancer cell death22.23
The present work is focused on designing, synthesizing, characterizing, and biologically assessing vanillin-isatin hybrids as potential MARK4 inhibitors. Our results demonstrated that the produced compounds had remarkable anticancer activities and were capable of binding to the active site of MARK4. Following the determination of binding affinity, the top-selected compounds were further explored for interaction analysis followed by cell-free and cell-based in vitro studies. Thus, current studies provided potential anticancer drug candidates targeting MARK4 in HCC.
2. Material and Methods
2.1. Materials and Instrumentation
All the chemicals and solvents utilized in the analytical procedures were obtained from Sigma-Aldrich (St. Louis, MO, USA). TLC aluminum sheets coated with F254 were purchased from Merck (India). Significant peaks in the IR spectra were captured by using an Agilent Cary 630 FT-IR spectrometer. Tetramethylsilane (TMS) was used as an internal standard to obtain 1H and 13C NMR spectra on Bruker Spectrospin DPX-300 spectrometer at 300 and 75 MHz, respectively. For splitting patterns, the letters s, d, t, and m and bars are used for singlet, doublet, triplet, multiplet, and broad singlet. Parts per million (ppm) values for 1H NMR chemical shifts (δ) are published about DMSO-d6, δ 2.54, whereas 13C NMR chemical shifts (δ) are recorded concerning DMSO-d6, 39.5. The mass spectra were recorded using an Agilent Quadrupole-6150, an LC/MS spectrometer. The digital Buchi melting point apparatus (M-560) was utilized to determine the uncorrected melting points. Using an X-bridge C18 having a 1.7m column with dimensions of 50 × 2.1 mm and an Agilent RRLC MS 6320 ion trap spectrometer, purities were determined. Ammonium acetate in water, 5 mM, comprises mobile phase channel A. The mobile phase B contained acetonitrile with a flow rate of 0.8 mL/min; 95% purity for each final compound was verified by using UV@214 detection. C3A (a subclone of the HepG2 cell line), SNU-475 (human live cancer cells), SH-SY5Y (human neuroblastoma cells), and HEK293 (human embryonic kidney cells) were taken from the National Centre for Cell Sciences (NCCS), Pune, India. The antitau (pSer262, cat:44–750 G), FITC labeled goat antirabbit IgG (31635) secondary antibodies, and dihydroethidium reagents were sourced from Invitrogen, Thermo Fisher Scientific. Furthermore, the FITC-Annexin-V detection kit was procured from BD-Pharmingen, BD Biosciences (USA). The reagents utilized in this study were of molecular biology grade.
2.2. Molecular Docking
We downloaded the coordinate file of MARK4 protein complexed with a cocrystallized ligand (PDB ID: 5ES1, 2.8 Å resolution) from the Protein Data Bank (http://www.rscb.org). Missing residues were remodeled through Modeler 10 in PyMOD3. Protein preparation was conducted using the Protein Preparation Wizard within Schrödinger Maestro 2022–4. The energy minimization of the protein structure was performed using the OPLS4 force field24−26 We utilized the LigPrep module within Schrodinger Maestro 2022–3 to prepare the VI-series ligands. We converted 2D structures to 3D structures, optimized geometry, and adjusted for chirality and desalting. Ionization and tautomeric states were determined using the Epik module, accommodating pH values ranging from 5 to 9. The libraries underwent minimization using the Optimized Potentials for Liquid Simulations-4 (OPLS-4) force field integrated with Schrodinger software. The libraries underwent minimization using the Optimized Potentials for Liquid Simulations-4 (OPLS-4) force field integrated within Schrodinger software.27 Subsequently, a single energetically favorable conformation was generated for each ligand, and these optimized ligands were utilized for docking analysis.26 Employing the site map tool of Maestro 2022–04 from of the Schrödinger suite, the binding site was defined to generate the grid. Subsequently, the prepared fragment libraries were docked against the binding pocket of the MARK4 protein using Glide and OPLS-4 force fields.28 The poses obtained after minimization underwent a rescoring procedure using the Glide Score scoring function. The docking process was performed with extra precision (XP)29−31
2.3. Molecular Dynamics Simulation
We utilized the Desmond module of Schrödinger Release 2022–4 to assess the protein binding interactions operating on a Linux system. The molecular dynamics (MD) simulation was performed to validate the structural integrity of the protein complex, employing the optimized potentials for liquid simulations (OPLS4) force field at pH 7.4. A 100 ns simulation was conducted to identify the optimal binding complexes. The process involved solvation of the chosen protein and the selected complex with water molecules within an orthorhombic box. To neutralize charges and maintain salt concentrations of 0.15M, Na+ and Cl– ions were added. Throughout the simulation, a consistent temperature of 300 K and a pressure of 1.01325 bar were maintained, with data recorded at intervals of 5 ps.32,33 The stability of the ligand-protein complex was evaluated by analyzing parameters such as the root-mean-square deviation (RMSD), root-mean-square fluctuation (RMSF), and the values of secondary structure elements (SSE). Furthermore, the interactions between the ligands and different atoms were analyzed for each trajectory frame. Additionally, the radius of gyration (Rg) was calculated to evaluate structural compression during the 100 ns simulation34−36 The Prime module from the Schrödinger suite 2022–4 was employed to calculate binding free energies between the ligand and receptor complex. The Molecular Mechanics-Generalized Born Surface Area (MM-GBSA) method, incorporating the OPLS4 force field, VSGB solvent model, and search algorithms, was utilized for this purpose.28
2.4. Protein Expression and Purification
Following our previously published procedures, MARK4 protein consisting of residues 59–368 was cloned, and, expressed in E. coli M15 cells and further purified.37
2.5. Fluorescence Binding Studies
Changes in the protein fluorescence intensity on a Jasco spectrofluorometer (Model FP-6200) with a protein concentration of 4 μM were used to track the binding of VI series (VI1–VI13) with MARK4. Experiment conditions included medium response, excitation wavelength of 280 nm, emission wavelength of 300–400 nm, and slit width of 10 nm. Following our previously described procedure,38 we were able to examine the binding affinities shown by the synthetic ester of the vanillin hydrazine isatin hybrid with recombinant MARK4. The number of binding sites (n) present on the protein and binding constant (Ka) were calculated using the modified Stern–Volmer equation based on a significant drop in protein fluorescence intensity with an increase in the concentration of ester of the vanillin-isatin hybrids. VI series compounds (7a to 7m) were used to titrate MARK4 at increasing concentrations (0–15 μM). Fluorescence emission data were fitted into the Stern–Volmer equation to derive the MARK4-VI series compound binding parameters.
2.6. Enzyme Inhibition Assay
The ATPase assay was employed to assess the enzyme activity of MARK4 in the presence of synthesized compounds and IC50 was calculated using the AAT Bioquest calculator as described previously.39,40 Briefly, to evaluate the activity of MARK4, a Malachite Green (Biomol Green reagent, Enzo Life Sciences) microtiter-plate assay was used. In the experimental setup, 6 μM MARK4 and 20 μM ATP were used as the substrate (in 20 mM Tris buffer, pH8.0.) and incubated at 25 °C for 10–15 min in the presence of various concentrations (ranging from 0 to 120 μM) of the synthesized ester of the vanillin hydrazine isatin hybrid. Following the incubation time, 100 μL of Biomol Green reagent was added to each well of the assay plate and the absorbance of reaction products were recorded at 620 nm using a BioRadmultiplate ELISA reader.
2.7. Cell Viability Studies
Human HCC cells SNU-475 and C3A (HepG2-subclone) were obtained from ATCC and cultured in Eagle’s Minimum Essential Medium (Corning, Cat # 10009-CVR) supplemented with 10% fetal bovine serum (Sigma Cat#12303C), and 1% penicillin + streptomycin solution (Invitrogen Cat#15,140–122). HEK293 (human noncancerous) cells were cultured in the RPMI 1640 medium (Gibco, Cat#11875119). A very sensitive cell viability reagent called PrestoBlue HS (resazurin-based high-sensitivity reagent, Invitrogen, cat. no. P50201) was used to conduct cell viability experiments on the selected cell lines. Briefly, a 96-well cell culture plate was seeded with 2000–3000 cells per well, and the plate was then incubated overnight to encourage growth. The next day, these cells were subjected to increasing doses ranging from 0 to 200 μM of MARK4 inhibitor molecules (compound VI-1 to VI-14). After 48–72 h of incubation of cells with compounds, 10 μL of PrestoBlue HS reagent was added to each well. The plates that had been treated were subsequently subjected to incubation with PrestoBlue reagent for approximately 45 min in a CO2 incubator. After the incubation period, a multiplate reader was used to measure the fluorescence of the plates using an excitation/emission filter set to 560/590 nm, respectively. The percentage cell viability was estimated to generate quantitative cell viability curves, taking vehicle control values as control, and plotting vs treatment doses of each compound as described previously.9,41
2.8. Tau-Phosphorylation Assay
In order to examine the inhibitory effect of the most active compound on MARK4, Tau-phosphorylation inhibition studies were performed as described.42 For this assay, we used human neuroblastoma cell line SH-SY5Y; these cells were maintained in a 1:1 mixture of Eagle’s minimum essential medium and F12 medium. These cells serve as model cell lines to study Tau phosphorylation. Briefly, the SH-SY5Y cells were added to a six-well cell culture plate and then incubated with VI-9 at its IC50 concentration (as obtained from enzyme inhibition studies). After a 24-h incubation period, the cells were harvested using trypsinization, subsequently fixed, rinsed, and then suspended in an antibody incubation buffer containing antiphosphatase primary antibodies and Alexa-647A conjugated secondary antibodies as described previously.42 Following the incubation, cells were washed to remove unbound/free antibodies, resuspended in FACS buffer, and analyzed by flow cytometry.
2.9. Transwell Cell Migration Assay
Transwell cell migration studies were performed to assess the effect of VI-9 treatment on the metastatic potentials of C3A and SNU-475 cells. Briefly, VI-9 was administered to the serum-starved cells at a concentration of 1/2 IC50 (the corresponding IC50 dosage for each cell line) for 24 h. After treatment, 50 × 103 cells/well of vehicle control or VI-9-treated cells were suspended in serum-free medium and placed in the upper chamber of the conventional Boyden chamber made of PET membranes (8.0 μm size, Cat#353093, Corning Life Sciences) as described previously.43 Conversely, 1 mL of complete media (including 10% FBS for chemotaxis) was placed in the lower chamber. After extracting the Boyden inserts on the following day, the migrating cells situated on the underside of the inset were immobilized using methanol and stained with crystal violet (0.4% in 25% methanol). The nonmigrated cells that were still present on the chamber’s top surface were removed using cotton swabs. Using ImageJ software (https://imagej.nih.gov/ij/), stained cells that had migrated to the bottom side of the Boyden chamber insert were imaged, quantified, and plotted for further analysis in four to five random fields for each condition.
2.10. Annexin-V/PI Staining
To evaluate the apoptosis induction potential of VI-9, annexin-V and propidium iodide (PI) staining studies were performed using flow cytometry as described previously.44,45 Briefly, compound VI-9 at a dose of 1/2 IC50 (derived from cell viability experiments) or vehicle control was administered to SNU-475 and C3A cells for 72 h. After being incubated with VI-9 for 72 h, the cells were trypsinized and washed 2–3 times with sterile PBS. Following the final PBS wash, the control and VI-9 treated cells were stained with PI (nuclear stain) and Cy5 labeled annexin-V antibodies (BD-Biosciences, San Jose, USA). The annexin-V/PI-stained cells were analyzed using flow cytometry. (FlowJo was used for analysis).
3. Results and Discussion
3.1. Chemistry
The vanillin-isatin hybrids were synthesized using a general synthetic approach, as shown in Scheme 1. Commercially available methyl 4-hydroxy-3-methoxybenzoate (1) was treated with hydrazine hydrate (2). The product obtained was subjected to nucleophilic substitution, and then condensation yielded vanillin hydrazide (3). A variety of aniline derivatives (4a-m) comprising both electron-withdrawing and electron-donating substituents underwent a series of reactions by reacting with chloral hydrate and hydroxylamine hydrochloride. As a result, iso-nitroso-acetanilide was obtained, whose cyclization resulted in the formation of substituted substituted isatins. Consequently, the substituted isatin derivatives were reacted with vanillin hydrazide (3) to form the corresponding vanillin-linked isatin hybrids (VI1–VI13). All the intermediates and title compounds, vanillin-isatin hydrazones, were characterized by multispectroscopic techniques such as FT-IR, 1H, 13C NMR, and mass spectrometry (Figures S1–S32).
Scheme 1. Reagents and Conditions: (a) Hydrazine Hydrate, Ethanol, Reflux at 80 °C; (b) Chloral Hydrate, NH2OH·HCl, Na2SO4, H2O, HCl (2N), 55 °C; (c) conc. H2SO4, 55-80°C; (d) Ethanol, Glacial Acetic Acid, Reflux at 80 °C for 8–12 h.
From the IR spectra, it was apparent that the formation of vanillin-isatin hydrazones (VI1–VI13), as demonstrated by the distinct peak at wave numbers 357–3400, 1667–1547, and 1696–1659 cm–1, which are indicative of the stretching motions of the NH, C=N, and C=O functional groups, respectively. O–H spectrum of vanillin shows a broad peak at 3268–3156 cm–1. The formation of vanillin-isatin hydrazone was also suggested by the presence of a sharp singlet in the range 13.93–13.87 ppm for the hydrazone proton −NH in the vanillin-isatin hydrazone 1H NMR spectra. Singlet observed in the range of 11.32–11.13 ppm is due to – NH proton of isatin. OH shows its characteristic singlet in the range of 10.10–10.03 ppm. The 13C NMR spectra revealed that the peaks attributed to isatin carbonyl functionality, and the carbonyl groups of hydrazones were exhibited in the ranges 164.22–163.59 and 159.89–151.75 ppm, respectively. The observed consistency between the mass spectra of all the compounds and the calculated values substantiated the successful synthesis of the targeted compounds. Finally, we confirmed that all the ultimate compounds had a minimum purity of ≥95%.
3.1.1. Synthesis of Hydrazide of Vanillin Ester (3)
In a solution of methyl 4-hydroxy-3-methoxybenzoate 1 (1.0 equiv) in ethanol (8 mL), hydrazine hydrate 2 (5.0 equiv) was added dropwise. The resulting mixture was subjected to reflux overnight at 80 °C. Formation of white precipitate is confirmed by TLC. Then, we filtered it and washed it with water; we obtained pure compound 3 and used it without further purification.
3.1.2. Synthesis of Substituted Isonitrosoacetanilides (5a-m)
The substituted isatins are well-known and reported earlier. Briefly the intermediate substituted isonitrosoacetanilides (5a-m) were synthesized as follows. First, various anilines (4a-m) (4.29 mmol) were introduced into a dry round-bottom flask for the reaction. Subsequently, water (23 mL) and 2 N of HCl (1.72 mL) were carefully added to the flask. The substituted aniline was completely dissolved before sequential addition of anhydrous sodium sulfate (28.35 mmol), hydroxylamine hydrochloride (15.03 mmol), and chloral hydrate (5.58 mmol) to the mixture. The reaction was carried out at 55 °C throughout the night, followed by cooling to room temperature upon completion, as confirmed by thin-layer chromatography (TLC). Afterward, the crude mixture was subjected to vacuum filtration and washed with distilled water, resulting in the formation of substituted isonitrosoacetanilides. These compounds were subsequently employed to synthesize isatin derivatives without further purification.
3.1.3. Synthesis of Substituted Isatins (6a-m)
In a round-bottom flask with two necks, concentrated sulfuric acid (1.08 mL) was gently heated to 50 °C, and subsequently, dry isonitrosoacetanilide (5a-m) (1.52 mmol) was introduced. The addition of the latter was carefully controlled to maintain the reaction vessel’s temperature within the range of 60–70 °C.46 To ensure the completion of the reaction, the temperature of the reaction vessel was further raised to 80 °C for 15–20 min, which was confirmed by TLC. Afterward, the reaction mixture was allowed to cool to room temperature and added to ice water in a 3-fold excess. The resulting solid residue was subjected to suction filtration, thoroughly washed with water to remove all traces of sulfuric acid, and finally dried to obtain high-purity substituted isatin (6a-m). All previously reported substitutions were considered.47
3.1.4. General Procedure for the Synthesis of Vanillin-Isatin Hydrazone Hybrids (VI1–VI13)
Vanillin hydrazide (3) (1 equiv) was properly dissolved in ethanol and added to an appropriate amount of isatin derivatives (6a-m) (1 equiv) in a dried RB flask. The mixture was then refluxed overnight at 80 °C after the addition of a few drops of glacial acetic acid. The reaction mixture was allowed to cool to room temperature following the completion of the reaction, and then ice-cold water was added. The filtrate was collected through a funnel, and ethanol was used to wash the resulting solid precipitate. Vacuum drying was employed to obtain the desired hybrids (VI1–VI13) from the resulting solid.
(Z)-4-Hydroxy-3-methoxy-N′-(2-oxoindolin-3-ylidene)benzohydrazide (VI-1) (7a)
Yellow solid, yield: 82.8%, Rf = 0.25 (ethyl acetate:hexane = 50:50), mp: 295.4 °C. IR (neat): ν (cm–1) 3514 (N–H)str. hydrazone, 3178 (O–H)broad., 1683 (C=O)str. hydrazone, and 1588 (C=N) isatin. 1H NMR (500 MHz, DMSO-d6) (δ, ppm): 13.88 (s, 1H, NHHydrazone), 11.32 (s, 1H, NHIsatine), 10.09 (s, 1H, OH), 7.57 (d, J = 5.0 Hz, 1H, Ar-H), 7.42 (d, J = 4.5 Hz, 1H, Ar-H), 7.34 (ddd, J = 1.0, 1.0, 2.1 Hz, 2H, Ar–H), 7.10–7.06 (m, 1H, Ar-H), 6.93 (dd, J = 8.0, 3.9 Hz, 2H, Ar-H), 3.82 (s, 3H, OCH3). 13C NMR (126 MHz, DMSO-d6) (δ-ppm):163.67 (C=O)Isatin, 151.81 (C=O)Hydrazone, 148.41, 142.74, 142.73, 132.10, 123.30, 123.28, 121.38, 120.45, 116.00, 111.90, 111.74, 56.15 (OCH3). ESI-MS (m/z) calcd. For C16H13N3O4: 311.30; Found: 310.08 [M-H]−.
(Z)-4-Hydroxy-3-methoxy-N′-(5-methyl-2-oxoindolin-3-ylidene)benzohydrazide (VI-2) (7b)
Dark yellow solid, yield: 92.3%, Rf = 0.22 (ethyl acetate:hexane = 50:50), mp: 294.2 °C. IR (neat): ν (cm–1) 3514 (N–H)str. hydrazone, 3219 (O–H)broad., 1670 (C = O)str.hydrazone, 1607 (C=N) isatin. 1H NMR (500 MHz, DMSO-d6) (δ, ppm):13.87 (s, 1H, NHHydrazone), 11.20 (s,1H, NHIsatine), 10.08 (s, 1H, OH), 7.41 (d, J = 1.9 Hz, 1H, Ar-H), 7.38 (s, 1H, Ar-H), 7.31 (dd, J = 2.1, 1.5 Hz, 1H, Ar-H), 7.14 (d, J = 7.9 Hz, 1H, Ar-H), 6.92 (d, J = 8.5 Hz, 1H, Ar-H), 6.81 (d, J = 7.9 Hz, 1H, Ar-H), 3.82 (s, 3H, OCH3), 2.26 (s, 3H, CH3). 13C NMR (126 MHz, DMSO-d6) (δ-ppm):163.72 (C=O)Isatin, 151.77 (C=O)Hydrazone, 148.39, 140.47, 132.49, 132.35, 123.33, 121.71, 121.36, 120.44, 115.98, 111.86, 111.49, 56.13(OCH3), 21.06 (CH3). ESI-MS (m/z) calcd. for C17H15N3O4: 325.32; Found: 324.13 [M-H]−.
(Z)-4-Hydroxy-3-methoxy-N′-(5-methoxy-2-oxoindolin-3-ylidene)benzohydrazide (VI-3) (7c)
Brick red solid, yield: 81.4%, Rf = 0.21 (ethyl acetate:hexane = 50:50), mp: 293.4 °C. IR (neat): ν (cm–1) 3409 (N–H)str. hydrazone, 3268 (O–H)broad., 1670 (C = O)str.hydrazone, 1570 (C=N) isatin. 1H NMR (500 MHz, DMSO-d6) (δ, ppm):13.93 (s, 1H, NHHydrazone), 11.13 (s, 1H, NHIsatine), 10.10 (s, 1H, OH), 7.42 (d, J = 1.9 Hz, 1H, Ar-H), 7.32 (dd, J = 2.1, 1.5 Hz, 1H, Ar-H), 7.10 (d, J = 1.5 Hz, 1H, Ar-H), 6.92 (d, J = 8.1 Hz, 2H, Ar-H), 6.84 (d, J = 8.5 Hz, 1H, Ar-H), 3.82 (s, 3H, OCH3), 3.74 (s, 3H, OCH3). 13C NMR (126 MHz, DMSO-d6) (δ-ppm):163.78 (C=O)Isatin, 155.95 (C=O)Hydrazone, 155.94, 151.83, 148.41, 136.39, 123.27, 121.45, 121.17, 118.42, 116.00, 112.61, 111.89, 106.11, 56.14 (OCH3), 56.12(OCH3). ESI-MS (m/z) calcd. for C17H15N3O5: 341.32; Found: 340.14 [M-H]−. HRMS: 342.1081[M+1]+.
(Z)-N′-(6-Chloro-2-oxoindolin-3-ylidene)-4-hydroxy-3-methoxybenzohydrazide (VI-4) (7d)
Yellow solid, yield: 84.6%, Rf = 0.25 (ethyl acetate:hexane = 50:50), mp: 314.5 °C. IR (neat): ν (cm–1) 3506 (N–H)str. hydrazone, 3164 (O–H)broad., 1670 (C = O)str.hydrazone, 1622 (C=N) isatin. 1H NMR (500 MHz, DMSO-d6) (δ, ppm):13.77 (s, 1H, NHHydrazone), 11.45 (s, 1H, NHIsatine), 10.11 (s, 1H, OH), 7.56 (d, J = 8.1 Hz, 1H, Ar-H), 7.41 (d, J = 2.0 Hz, 1H, Ar-H), 7.31 (dd, J = 2.1, 2.0 Hz, 1H, Ar-H), 7.10 (dd, J = 2.1, 1.5 Hz, 2H, Ar-H), 6.94 (d, J = 1.9 Hz, 1H, Ar-H), 3.82 (s, 3H, OCH3). 13C NMR (126 MHz, DMSO-d6) (δ-ppm):163.64(C = O)Isatin, 151.89(C = O)Hydrazone, 151.48, 148.41, 147.93, 143.86, 136.06, 123.15, 122.72, 122.09, 119.40, 116.00, 115.56, 111.76, 111.12, 56.13 (OCH3). ESI-MS (m/z) calcd. for C16H12ClN3O4: 345.74; Found: 344 [M-H]−.
(Z)-N′-(5,7-Dimethyl-2-oxoindolin-3-ylidene)-4-hydroxy-3-methoxybenzohydrazide (VI-5) (7e)
Dark yellow solid, yield: 90.6%, Rf = 0.21 (ethyl acetate:hexane = 50:50), mp: 315.4 °C. IR (neat): ν (cm–1) 3400 (N–H)str. hydrazone, 3156 (O–H)broad., 1666 (C=O)str.hydrazone, 1588 (C=N) isatin. 1H NMR (500 MHz, DMSO-d6) (δ, ppm):13.90 (s, 1H, NHHydrazone), 11.23 (s, 1H, NHIsatine), 10.08 (s, 1H, OH), 7.42 (d, J = 2.0 Hz, 1H, Ar-H), 7.32 (dd, J = 2.1, 1.9 Hz, 1H, Ar-H), 7.22 (s, 1H, Ar-H), 6.98 (s, 1H, Ar-H), 6.92 (d, J = 8.1 Hz, 1H, Ar-H), 3.82 (s, 3H, OCH3), 2.23 (s, 3H, CH3), 2.15 (d, J = 7.4 Hz, 3H, CH3). 13C NMR (126 MHz, DMSO-d6) (δ-ppm):164.15 (C=O)Isatin, 151.75 (C=O)Hydrazone, 151.11, 151.04, 148.41, 146.31, 139.13, 133.94, 132.28, 123.37, 120.88, 120.19, 119.13, 115.98, 56.13 (OCH3), 20.97 (CH3), 16.38 (CH3). ESI-MS (m/z) calcd. for C18H17N3O4: 339.35; Found: 338 [M-H]−.
(Z)-N′-(5-Chloro-2-oxoindolin-3-ylidene)-4-hydroxy-3-methoxybenzohydrazide (VI-6) (7f)
Yellow solid, yield: 85.6%, Rf = 0.24 (ethyl acetate:hexane = 50:50), mp: 314.6 °C. IR (neat): ν (cm–1) 3499 (N–H)str. hydrazone, 3216 (O–H)broad., 1674 (C = O)str.hydrazone, 1603 (C=N) isatin. 1H NMR (500 MHz, DMSO-d6) (δ, ppm):13.83 (s, 1H, NHHydrazone), 11.44 (s, 1H, NHIsatine), 10.13 (s, 1H, OH), 7.57 (d, J = 5.1 Hz, 1H, Ar-H), 7.43–7.38 (m, 2H, Ar-H), 7.33 (dd, J = 2.5, 2.0 Hz, 1H, Ar-H), 6.95–6.92 (m, 2H, Ar-H), 3.82 (s, 3H, OCH3). 13C NMR (126 MHz, DMSO-d6) (δ-ppm):151.97 (C=O) Hydrazone, 150.96, 148.44, 144.78, 141.41, 127.42, 123.10, 122.21, 120.90, 116.04, 56.15 (OCH3). ESI-MS (m/z) calcd. for C16H12ClN3O4: 345.74; Found: 343 [M-2H]−.
(Z)-N′-(5-Bromo-2-oxoindolin-3-ylidene)-4-hydroxy-3-methoxybenzohydrazide (VI-7) (7g)
Yellow solid, yield: 82.5%, Rf = 0.25 (ethyl acetate:hexane = 50:50), mp: 313.2 °C. IR (neat): ν (cm–1) 3502 (N–H)str. hydrazone, 3231 (O–H)broad., 1674 (C=O)str.hydrazone, 1603 (C=N) isatin. 1H NMR (500 MHz, DMSO-d6) (δ, ppm):13.82 (s, 1H, NHHydrazone), 11.45 (s, 1H, NHIsatine), 10.13 (s, 1H, OH), 7.78–7.29 (m, 4H, Ar-H), 6.92–6.89 (m, 2H, Ar-H), 3.82 (s, 3H, OCH3). 13C NMR (126 MHz, DMSO-d6) (δ-ppm):153.65 (C=O)Hydrazone, 145.60, 141.12, 133.36, 126.10, 125.51, 121.63, 119.24, 114.16, 112.37, 109.69, 56.09 (OCH3). ESI-MS (m/z) calcd. for C16H12BrN3O4: 390.19; Found: 388 [M-2H]−.
(Z)-4-Hydroxy-3-methoxy-N′-(7-nitro-2-oxoindolin-3-ylidene)benzohydrazide (VI-8) (7h)
Yellow solid, yield: 87.2%, Rf = 0.26 (ethyl acetate:hexane = 50:50), mp: 319.6 °C. IR (neat): ν (cm–1) 3512 (N–H)str. hydrazone, 3198 (O–H)broad., 1687 (C=O)str.hydrazone, 1599 (C=N) isatin. 1H NMR (500 MHz, DMSO-d6) (δ, ppm):13.73 (s, 1H, NHHydrazone), 11.95 (s, 1H,NHIsatine), 10.12 (s, 1H, OH), 8.15–7.97 (m, 2H, Ar-H), 7.45–7.27 (m, 3H, Ar-H), 6.95 (d, J = 8.1 Hz, 1H, Ar-H), 3.84 (s, 3H, OCH3). 13C NMR (126 MHz, DMSO-d6) (δ-ppm):164.21(C=O)Isatin, 152.12 (C=O)Hydrazone, 148.48, 132.44, 130.91, 126.98, 126.08, 123.27, 116.02, 115.66, 56.04. (OCH3). ESI-MS (m/z) calcd. for C16H12N4O6: 356.29; Found: 355 [M-H]−.
(Z)-N′-(7-Chloro-2-oxoindolin-3-ylidene)-4-hydroxy-3-methoxybenzohydrazide (VI-9) (7i)
Yellow solid, yield: 88.2%, Rf = 0.22 (ethyl acetate:hexane = 50:50), mp: 322.7 °C. IR (neat): ν (cm–1) 3517 (N–H)str. hydrazone, 3227 (O–H)broad., 1681 (C=O)str.hydrazone, 1547 (C=N) isatin. 1H NMR (500 MHz, DMSO-d6) (δ, ppm):13.81 (s, 1H, NHhydrazone), 11.75 (s, 1H, NHIsatine), 10.13 (s, 1H, OH), 7.54 (d, J = 7.5 Hz, 1H, Ar-H), 7.42 (d, J = 4.5 Hz, 2H, Ar-H), 7.34 (d, J = 9.5 Hz, 1H, Ar-H), 7.11–6.92 (m, 2H, Ar-H), 3.82 (s, 3H, OCH3). 13C NMR (126 MHz, DMSO-d6) (δ-ppm):163.75 (C=O)Isatin, 151.97 (C=O) Hydrazone, 148.53, 140.03, 131.57, 124.39, 123.11, 122.50, 119.92, 116.03, 115.80, 111.97, 56.10 (OCH3). ESI-MS (m/z) calcd. for C16H12ClN3O4: 345.74; Found: 344 [M-H]−.
(Z)-N′-(6-Chloro-5-fluoro-2-oxoindolin-3-ylidene)-4-hydroxy-3-methoxybenzohydrazide (VI-10) (7j)
Yellow solid, yield: 91.3%, Rf = 0.21 (ethyl acetate:hexane = 50:50), mp: 346.4 °C. IR (neat): ν (cm–1) 3514 (N–H)str. hydrazone, 3178 (O–H)broad., 1670 (C = O)str.hydrazone, 1603 (C=N) isatin. 1H NMR (500 MHz, DMSO-d6) (δ, ppm):14.01 (s, 1H,NHHydrazone), 11.52 (s, 1H, NHIsatine), 10.13 (s, 1H, OH)), 7.45–7.30 (m, 3H, Ar-H), 6.97–6.84 (m, 2H,Ar-H), 3.82 (s, 3H, OCH3). 13C NMR (126 MHz, DMSO-d6) (δ-ppm):163.59 (C=O)Isatin, 153.34 (C=O) Hydrazone, 151.96, 148.30, 139.64, 118.56, 116.13, 110.84, 56.14(OCH3). ESI-MS (m/z) calcd. for C16H11ClFN3O4: 363.73; Found: 362 [M-H]−.
(Z)-N′-(6-Fluoro-2-oxoindolin-3-ylidene)-4-hydroxy-3-methoxybenzohydrazide (VI-11) (7k)
Dark yellow solid, yield: 85.3%, Rf = 0.26 (ethyl acetate:hexane = 50:50), mp: 317.5 °C. IR (neat): ν (cm–1) 3510 (N–H)str. hydrazone, 3164 (O–H)broad., 1696 (C=O)str.hydrazone, 1603 (C=N) isatin. 1H NMR (500 MHz, DMSO-d6) (δ, ppm):13.88 (s, 1H, NHHydrazone), 11.34 (s, 1H, NHIsatine), 10.12 (s, 1H, OH), 7.44–7.30 (m, 3H, Ar-H), 7.19 (td, J = 9.2, 2.6 Hz, 1H, Ar-H), 6.92 (d, J = 8.2 Hz, 2H, Ar-H), 3.82 (s, 3H, OCH3). 13C NMR (126 MHz, DMSO-d6) (δ-ppm):163.84 (C=O)Isatin, 159.83 (C=O)Hydrazone, 158.10, 151.94, 148.43, 138.98, 138.97, 123.12, 118.50, 116.01, 112.80, 111.91, 108.56, 108.35, 56.09 (OCH3). ESI-MS (m/z) calcd. For C16H12FN3O4: 329.29; Found: 328 [M-H]−.
(Z)-4-Hydroxy-3-methoxy-N′-(6-methoxy-2-oxoindolin-3-ylidene)benzohydrazide (VI-13) (7l)
Brown solid, yield: 88.5%, Rf = 0.25 (ethyl acetate:hexane = 50:50), mp: 360.2 °C. IR (neat): ν (cm–1) 3514 (N–H)str. hydrazone, 3212 (O–H)broad., 1659 (C=O)str.hydrazone, 1603 (C=N) isatin. 1H NMR (500 MHz, DMSO-d6) (δ, ppm):13.91 (s, 1H, NHHydrazone), 11.12 (s, 1H, NHIsatine), 10.24 (s, 1H, OH), 7.41 (d, J = 1.7 Hz, 1H, Ar-H), 7.33 (dd, J = 8.1, 1.8 Hz, 1H, Ar-H), 7.11 (d, J = 1.3 Hz, 1H, Ar-H), 6.86 (d, J = 8.2 Hz, 2H, Ar-H), 6.81 (d, J = 8.5 Hz, 1H, Ar-H), 3.8111 (s, 3H, OCH3), 3.74 (s, 3H, OCH3). 13C NMR (126 MHz, DMSO-d6) (δ-ppm):163.77 (C=O)Isatin, 155.85 (C=O)Hydrazone, 155.91, 151.82, 148.41, 136.33, 123.22, 121.41, 121.11, 118.42, 116.10, 112.51, 111.83, 56.14 (OCH3), 56.12 (OCH3). ESI-MS (m/z) calcd. For C17H15N3O5: 341.32; Found: 340 [M-H]−.
(Z)-N′-(6-Fluoro-2-oxoindolin-3-ylidene)-4-hydroxy-3-methoxybenzohydrazide (VI-14) (7m)
Yellow solid, yield: 92.1%, Rf = 0.22 (ethyl acetate hexane = 50:50), mp: 308.9 °C. IR (neat): ν (cm–1) 3517 (N–H)str. hydrazone, 3164 (O–H)broad., 1670 (C=O)str.hydrazone, 1614 (C=N) isatin. 1H NMR (500 MHz, DMSO-d6) (δ, ppm):13.75 (s, 1H, NHHydrazone), 11.45 (s, 1H, NHIsatine), 10.04 (s, 1H, OH), 7.59 (dd, J = 6.1, 3.0, Hz, 1H,Ar-H), 7.41–7.31(m, 2H, Ar-H), 6.92–6.74 (m, 3H, Ar-H), 3.82 (s, 3H, OCH3). 13C NMR (126 MHz, DMSO-d6) (δ-ppm):163.91 (C=O)Isatin, 151.85 (C=O)Hydrazone, 148.40, 144.51, 144.41, 123.31, 116.91, 116.00, 111.90, 110.01, 109.86, 100.01, 99.85, 56.0 (OCH3). ESI-MS (m/z) calcd. For C16H12FN3O4: 329.29; Found: 328 [M-H]−.
3.2. Molecular Docking Studies
In previous studies, isatin-triazole hydrazones were found to show promising anticancerous activity and as potential inhibitors of MARK4.48 Molecular docking analysis is used to calculate the binding affinity and the interacting MARK4 residues with the ligand, and the outcomes are presented in (Table 1 and Figure 2). The best-docked complexes were VI-9, according to the interacting residues and binding energy determined by molecular docking studies.
Table 1. Docking Parameters of VI Series with MARK4.
Figure 2.
Molecular docking: (A) Ribbon structure showing interaction of vanillin-isatin hybrids (VI-4, VI-5, VI-8, and VI-9) complexed with MARK4 protein. (B) Interacting residues of MARK4 with vanillin-isatin hybrids (VI-4, VI-5, VI-8, and VI-9). (C) Focused view MARK4 active site complexed with vanillin-isatin hybrids (VI-4, VI-5, VI-8, and VI-9).
Most of the vanillin-isatin hybrids bind to MARK4 and showed strong interactions to Ile62, Val70, Ala83, Lys85, Tyr134, Ala135, Leu185, Ala195 and Asp196. The interaction between VI-9 and MARK4 is facilitated by forming hydrogen bonds with Lys85 (Figure 2A). In addition to its potent hydrogen bonding, VI-9 generates diverse interactions with Ile62, Val70, Ala83, Leu185, and Ala195 (Figure 2). Ester of vanillin forms a hydrogen bond with MARK4. This bond is between Ala135 and the hydroxyl group of the vanillin ester (Figures S33 and S34). The ester of vanillin has been observed to interact with additional amino acid residues of MARK4, namely, Ile62, Val70, Ala83, and Leu185.
The Glide module of Schrödinger suite 2022–4 was employed for extra precision docking (XP) within the predicted catalytic pocket of MARK4. The ligand’s docking score obtained through XP-docking was −7.877 kcal/mol. The Binding Free Energy (BFE) value was calculated to be −61.00 kcal/mol. Notably, the hydroxyl group of 4-Hydroxy-3-methoxy Benz amide moiety of the ligand formed two hydrogen bonds with Glu133 and Ala135. In addition, Asn183 formed a halogen bond with the chloride of the ligand (Figure 2B).
3.3. MD Simulation Studies
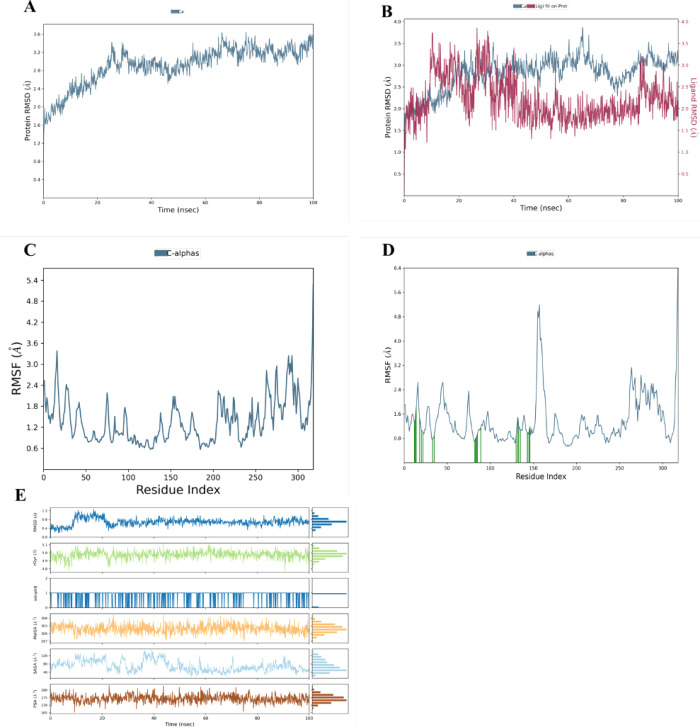
MD simulation was employed to investigate the dynamic properties of protein–ligand complexes at the atomic level. The stability of the protein and ligands was assessed during a 100 ns MD simulation using the apoprotein and the DHPS-8F complex as references. The RMSD was calculated as 2.0 to 3.6 Å, with a gradual initial increase in the first 20 ns from 2.0 to 2.5 Å, followed by a rapid increase to reach 3.5 Å in 10 ns only (20–30 ns), and after that, the system underwent subsequent stabilization. The RMSD of the MARK4-VI9 complex did not show a significant difference in values compared to the apoprotein except for the sharp and sudden fluctuations exhibited by the complex, which happened for a very short time and were in the accepted ranges. The range of RMSD values of the complex indicates stronger binding.
RMSF was calculated for all complexes to assess residue flexibility, revealing that the residues that formed H-bonds with the protein had very low RMSD values, such as Glu-133, Ala-135, and Asp-196, which showed RMSF values as low as 0.80, 0.94, and 1.19 Å respectively. Furthermore, the RMSF value of Glu-139, which formed a significant water bridge with VI-9 for more than 90% of the simulation time, was 1.16 Å. Thus, the protein’s binding pocket is relatively stable. The ligands’ compactness was evaluated through Rg values ranging from 4.8 to 5.1 Å; these results show that VI-9 possesses good structural rigidity. SASA analysis shows that the ligand has a meager SASA value ranging from 40 to 120 Å2, indicating that a substantial portion of VI-9 is correctly situated in the protein binding pocket (Figure 3A–E).
Figure 3.
Structural dynamics of MARK4 following binding of VI-9. (A) RMSD plot of MARK4 protein. (B) RMSD plot of MARK4 complex with VI-9. (C) Average residual fluctuations of the MARK4 protein. (D) Average residual fluctuations plot of the MARK4 protein complex with VI-9. (E) Radius of gyration (Rg) and SASA plots of ligand VI-9.
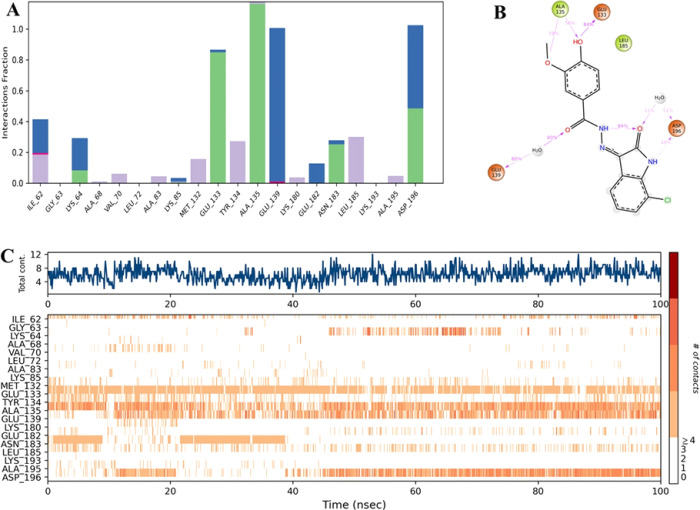
Several hydrogen bonds were observed to form between VI-9 and specific residues, such as Glu-133, Ala-135, and Asp-196. These H-bonds lasted 100%, 85%, and 50% of the simulation time, respectively. Strong water bridge interactions were reported with Glu-139 and Asp-196 residues, which lasted 100% and 50% of the simulation time, respectively. Detailed insights into these interactions are provided in Figure 4.
Figure 4.
Analysis of the interactions between the ligand and the protein’s backbone. (A) Showing hydrogen bonds between VI-9 and specific residues. (B) Representation of detailed ligand atom interactions with the protein residues. (C) Timeline representation of the interactions and contacts (H-bonds, Hydrophobic, Ionic, Water bridges).
3.4. Fluorescence Binding Studies
We measured the binding constants and the number of binding sites of ligands to MARK4 using fluorescence spectroscopy to validate the docking results. MARK4 protein was expressed and purified which showed a single band on SDS-PAGE was taken for this study (Figure S35).49 We titrated MARK4 (5 μM) by adding 1 μM selected synthetic compounds in each titration. MARK4 was excited at a wavelength of 280 nm and subsequent emission spectra were recorded in the 300–400 nm. Compounds with high binding affinities exhibit saturation at low concentrations, whereas those with weak affinities remain unbound even at high concentrations (12 μM). For clarity and straightforward interpretation of results, we provided the refined spectrum of fluorescence binding studies, which showed the quenching in the fluorescence emission spectrum of MARK4 with the increasing concentrations of ligand (Figure 5A). The results of the fluorescence were consistent with those of the docking studies.
Figure 5.
Binding studies of VI-9 with MARK4. (A) Fluorescence emission spectra of MARK4 (4 μM) were recorded with increasing concentrations of VI-9 (0–15 μM). (B) Stern–Volmer plot generated by quenching MARK4 fluorescence with increasing concentrations of VI-9. (C) Modified Stern–Volmer plot was acquired through the quenching experiments of the fluorescence of MARK4, introducing different concentrations of VI-9.
The Stern–Volmer equation shows the inverse connection between each synthetic derivative’s concentration and fluorescence intensity. However, fluorescence quenching data was analyzed with the help of the modified Stern–Volmer equation (MSV) to estimate various binding parameters, such as Ka and n, representing the binding constant and number of binding sites per protein molecule. VI-9 and VI-13, which only have one binding site, have binding affinities of 1.48 × 105 M–1 and 2.67 × 107 M–1, respectively (Table 2). Ka was estimated for VI-9 to be 1.48 × 105 M–1 (Figure 5C), indicating a good binding as well. However, another examined ester of the vanillin hydrazine isatin hybrid did not demonstrate any discernible binding. VI-9 and VI-13 appear to be the most active derivatives against MARK4 among all synthesized compounds (Table 2).
Table 2. Binding Properties of Vanillin-Isatin Hybrids with the MARK4.
| S. No. | Code | Ka (M–1) | n | KSV (M–1) |
|---|---|---|---|---|
| 1 | VI-1 | 5.6 × 106 | 1.241 | 3.9 × 105 |
| 2 | VI-2 | 1.38 × 106 | 1.145 | 3.7 × 105 |
| 3 | VI-3 | 2.25 × 107 | 1.367 | 3.8 × 105 |
| 4 | VI-4 | 8.96 × 105 | 1.141 | 1.8 × 105 |
| 5 | VI-5 | 2.5 × 104 | 0.8404 | 1.0 × 105 |
| 6 | VI-6 | 6.38 × 105 | 1.135 | 1.4 × 105 |
| 7 | VI-7 | 3.32 × 105 | 1.067 | 1.7 × 105 |
| 8 | VI-8 | 1.31 × 104 | 0.8606 | 0.6 × 105 |
| 9 | VI-9 | 1.48 × 105 | 1.047 | 1.0 × 105 |
| 10 | VI-10 | 7.9 × 104 | 1.006 | 0.8 × 105 |
| 11 | VI-11 | 1.56 × 105 | 1.015 | 1.4 × 105 |
| 12 | VI-13 | 2.67 × 107 | 1.517 | 0.8 × 105 |
| 13 | VI-14 | 1.36 × 106 | 1.199 | 1.8 × 105 |
3.5. Enzyme Inhibition Studies
Earlier, we reported that phytochemicals significantly affect MARK4 activity.9,42,50,51 To further optimize the MARK4-specific activity profile of vanillin, we synthesized a series of vanillin ester hydrazone-based isatin-hybrid molecules. We evaluated them with MARK4 using binding studies and other activity assays (Figure 6). All of the synthesized vanillin–isatin hybrids were tested for MARK4 inhibition using an ATPase inhibition assay. The ATPase activity of MARK4 decreased considerably in the presence of VI-2, VI-3, VI-5, and VI-9 (Figure 6). Furthermore, it was observed that VI-9 acts as the most prominent inhibitor for MARK4, as evidenced by the lowest IC50 dose of 7.16 μM. The outcomes of enzyme inhibition studies suggest that VI-9 inhibits MARK4 with a high affinity; therefore, it was selected for further biological evaluations.
Figure 6.
Enzyme inhibition assay of VI-2, VI-3, VI-5, and VI-9 and their IC50 values (A–D).
3.6. Cell-Based Assays
MARK4 is a known target for various cancer types, including HCC.50,52 We performed binding studies of these molecules with recombinant MARK4 for initial screening and further screened them against HCC cell lines. Interestingly, these molecules showed differential anticancer profiles toward SNU-475 and C3A cell lines of HCC (Figure 7A, B). The selected compounds decreased the viability of SNU-475 and C3A cell lines in a dose-dependent manner (Figure 7A, B). We have selected the most active compound, VI-9, by comparing its IC50 values with all the synthesized compounds. The IC50 value of VI-9 on SNU-475 and C3A cells was 4.35 μM and 6.96 μM, respectively (Figure 7C). These results also correlated with the outcomes of binding and enzyme inhibition studies; therefore, we selected VI-9 for further studies and validated its MARK4-specific activity profile through tau phosphorylation studies in neuroblastoma cell lines (SH-SY5Y), as these cell lines were considered as model cells to study tau-related pathologies.9,53 Interestingly, it was observed that the treatment of VI-9 decreased the tau phosphorylation in SH-SY5Y cells (Figure 7D and Figure S36). As tau is a substrate for MARK4 (MARK4 phosphorylates tau), a decrease in the phosphorylation of tau suggested that VI-9 treatment inhibited the activity of MARK4. This indicates that VI-9 possesses MARK4-specific activities.
Figure 7.
Cell viability studies and tau phosphorylation. Cell viability studies of all synthesized compounds following 72 h treatment on (A) SNU-475 cell line and (B) C3A cell line. (C) Cell viability plot for the most active VI-9 compound on SNU-475 and C3A cell lines. (D) The tau phosphorylation studies of VI-9 on SH-SY5Y neuroblastoma cells. Following the treatment of the cells with vehicle control/VI-9, they were fixed, stained with anti-p-Tau antibodies, and analyzed through flow cytometry.
MARK4 is also involved in cancer cell migration;7 therefore, we have studied the effect of VI-9 treatment on the cell migration properties of HCC cells. We treated the cells with VI-9 and performed a transwell migration assay using Boyden chamber inserts. Compared to the vehicle control, it was observed that VI-9 treatment significantly decreased the migration of SNU-475 and C3A cells (Figure 8A, B). Moreover, up to 200 μM concentration, VI-9 did not exhibit cytotoxicity or detrimental effects on noncancerous HEK293 cells (Figure S37).
Figure 8.
Cell migration (metastasis) and apoptosis studies on HCC cells. (A) Representative images illustrate the results of transwell cell migration experiments involving vehicle control/VI-9 administration to SNU-475 and C3A cells. (B) The quantification of migrated cells in the presence of vehicle control compared to the treatment with VI-9 in C3A and SNU-475 cell lines. (C) Scattered dot-plot with heat map representation for annexin-V/PI staining from vehicle control or VI-9 treated SNU-475 and C3A cells, as accessed using flow cytometry. (D) The quantification of annexin-V/PI positive cells in vehicle control vs VI-9 treated HCC cell lines. A t test was employed to determine the significance of the analysis (from n = 3 biological replicates).
Additionally, the apoptotic potential of VI-9 was studied on SNU-475 and C3A cells through a flow-cytometry-based annexin-V/PI staining assay. Consequently, VI-9 has been shown to strongly promote early apoptosis in SNU-475 and C3A cell lines (Figure 8C, D). These outcomes showed that VI-9 is a promising anticancer molecule targeting MARK4. All of the cell-based functional studies of VI-9 on HCC cells collectively point to a substantial level of anticancer activity with a MARK4-specific profile. These findings support the potential utilization of analogs derived from isatin in cancer therapy by targeting MARK4. Certain molecules VI-9 demonstrated significant antiproliferative effects against liver cancer cell lines, with higher docking scores than other compounds, indicating their promising efficacy.
Indeed, targeted therapeutic medications are now often used to treat cancer because of their benefits over conventional chemotherapy medications in terms of effectiveness, specificity, and safety. Small-molecule inhibitors are made to specifically target proteins or enzymes involved in the initiation and spread of cancer. By inhibiting they perturb signaling pathways that foster growth, metastasis, and resistance to cancer treatment. Small molecule inhibitor-based targeted therapeutic approaches are the subject of much research attention. Due to their small size, they can interact with the cytoplasmic domain of intracellular signaling molecules and cell-surface receptors via translocation through the plasma membrane. Most of the small molecule inhibitors block essential targets for cancer including proteosomes, heat shock proteins, serine/threonine/tyrosine kinases, and other proteins involved in signal transduction pathways. Finding specific genes or proteins and comprehending the mechanism(s) driving the development of each cancer can aid in developing creative approaches to increase the effectiveness of existing therapies and perhaps even lead to the discovery of new therapies.
4. Conclusions
The present study summarizes that esters of vanillin and their hydrazine isatin hybrids significantly bind to the active site cavity of MARK4 and inhibit its kinase activity. Among all synthesized compounds, VI-9 exhibited the most promising inhibitory effect against MARK4. The suppression of MARK4 activity presumably hampers the growth of the cancerous cells. The outcomes derived from our preclinical studies strengthen our hypothesis and correlate with the previous studies,54,55 affirming the potential of MARK4 as a potential target for cancer therapy. The selected lead molecule VI-9 also shows specificity toward MARK4, and it was observed that the level of binding affinity displayed by the esters of vanillin hydrazine isatin hybrid toward the active site cavity of MARK4, which could potentially aid in enzyme inhibition. Moreover, our findings highlight the potential of an ester of vanillin and its vanillin-isatin hybrids as promising candidates for developing MARK4 inhibitors with potent anticancer properties.
Acknowledgments
S.A. acknowledges that the Non-Net Fellowship is the financial support from the University Grant Commission (UGC), Government of India. MIH is thankful to the Central Council for Research in Unani Medicine (CCRUM) Ministry of Ayush, Government of India (ref No. F. No.3–69/2020-CCRUM/Tech). A.H. acknowledges the generous support from the researchers supporting project number RSPD2024R980, King Saud University, Riyadh, Saudi Arabia. II acknowledges DST for postdoctoral fellowship under the women scientist scheme (File no: SR/WOS-A/CS-116/2018).
Data Availability Statement
All data analyzed during this study are included in this manuscript and the Supporting Information attached to this paper.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.4c00661.
1HNMR, 13C NMR, and mass spectra and UP-LC purity analyses of VI-1–VI-9 and VI-14; 3D views and 2D interaction diagrams; analysis of binding interactions using molecular docking studies; SDS-PAGE of purified MARK4 protein; cytotoxicity studies of VI-9 on HEK293 cells; tau phosphorylation studies of VI-9 on SH-SY5Y cells. (PDF)
Author Contributions
Sarfraz Ahmed: Conceptualization, formal analysis, writing-original draft; Aarfa Queen: Visualization, formal analysis, Data Analysis, writing-original draft; Iram Irfan: Data validation, Data Analysis; writing-review and editing; Mohammad Naseem Siddiqui: Data analysis, Editing, Data curation; Haider Thaer Abdulhameed Almuqdadi: Visualization, software, Data validation, Writing-review and editing; Nisha Setia: writing-review and editing; Jaoud Ansari: Data validation, data curation; Afzal Hussain: data curation, software Writing-review and editing; Md. Imtaiyaz Hassan: Conceptualization, Data curation, Supervision, Writing-review and editing, formal analysis, Project Administration. Mohammad Abid: Conceptualization, Data curation, Supervision, Data validation, Writing-review and editing, and Project Administration. All authors reviewed and approved the final version of the manuscript.
This work is supported by the Central Council for Research in Unani Medicine (CCRUM) Ministry of Ayush, Government of India (ref No. F.No.3–69/2020-CCRUM/Tech) awarded to M.I.H., and Researchers supporting project (RSPD2024R980) King Saud University, awarded to A.H.
The authors declare no competing financial interest.
Supplementary Material
References
- Drewes G.; Ebneth A.; Mandelkow E.-M. MAPs, MARKs and microtubule dynamics. Trends in biochemical sciences. 1998, 23 (8), 307–311. 10.1016/S0968-0004(98)01245-6. [DOI] [PubMed] [Google Scholar]
- Trinczek B.; Brajenovic M.; Ebneth A.; Drewes G. MARK4 is a novel microtubule-associated proteins/microtubule affinity-regulating kinase that binds to the cellular microtubule network and to centrosomes. J. Biol. Chem. 2004, 279 (7), 5915–5923. 10.1074/jbc.M304528200. [DOI] [PubMed] [Google Scholar]
- Ahmed S.; Mobashir M.; Al-Keridis L. A.; et al. A Network-Guided Approach to Discover Phytochemical-Based Anticancer Therapy: Targeting MARK4 for Hepatocellular Carcinoma. Front Oncol. 2022, 12, 914032. 10.3389/fonc.2022.914032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnani I.; Novielli C.; Fontana L.; et al. Differential signature of the centrosomal MARK4 isoforms in glioma. Analytical Cellular Pathology. 2011, 34 (6), 319–338. 10.1155/2011/206756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W.; Lee S.; Huang X.; Liu S.; Inayathullah M.; Kim K.-M.; Tang H.; Ashford J. W.; Rajadas J.; et al. Attenuation of synaptic toxicity and MARK4/PAR1-mediated Tau phosphorylation by methylene blue for Alzheimer’s disease treatment. Scientific reports. 2016, 6 (1), 34784. 10.1038/srep34784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato T.; Satoh S.; Okabe H.; et al. Isolation of a novel human gene, MARKLI, homologous to MARK3 and its involvement in hepatocellular carcinogenesis. Neoplasia. 2001, 3 (1), 4–9. 10.1038/sj.neo.7900132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidary Arash E.; Shiban A.; Song S.; Attisano L. MARK4 inhibits Hippo signaling to promote proliferation and migration of breast cancer cells. EMBO reports. 2017, 18 (3), 420–436. 10.15252/embr.201642455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beghini A.; Magnani I.; Roversi G.; et al. The neural progenitor-restricted isoform of the MARK4 gene in 19q13. 2 is upregulated in human gliomas and overexpressed in a subset of glioblastoma cell lines. Oncogene. 2003, 22 (17), 2581–2591. 10.1038/sj.onc.1206336. [DOI] [PubMed] [Google Scholar]
- Khan P.; Rahman S.; Queen A.; Manzoor S.; Naz F.; Hasan G. M.; Luqman S.; Kim J.; Islam A.; Ahmad F.; Hassan M. I. Elucidation of dietary polyphenolics as potential inhibitor of microtubule affinity regulating kinase 4: in silico and in vitro studies. Scientific reports. 2017, 7 (1), 9470. 10.1038/s41598-017-09941-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naz F.; Khan F. I.; Mohammad T.; et al. Investigation of molecular mechanism of recognition between citral and MARK4: A newer therapeutic approach to attenuate cancer cell progression. International journal of biological macromolecules. 2018, 107, 2580–2589. 10.1016/j.ijbiomac.2017.10.143. [DOI] [PubMed] [Google Scholar]
- Sumpter W. C. The chemistry of isatin. Chemical reviews. 1944, 34 (3), 393–434. 10.1021/cr60109a003. [DOI] [Google Scholar]
- Ajin P. K.Treatment of Depression in Rats Exposed To Chronic Unpredictable Mild Stress Using Synthetic Derivatives of Isatin; Sri Ramakrishna Institute of Paramedical Sciences: Coimbatore, 2020. [Google Scholar]
- Ferraz de Paiva R. E.; Vieira E. G.; Rodrigues da Silva D.; Wegermann C. A.; Costa Ferreira A. M. Anticancer compounds based on isatin-derivatives: Strategies to ameliorate selectivity and efficiency. Frontiers in molecular biosciences. 2021, 7, 627272. 10.3389/fmolb.2020.627272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendel D. B.; Laird A. D.; Xin X.; et al. In vivo antitumor activity of SU11248, a novel tyrosine kinase inhibitor targeting vascular endothelial growth factor and platelet-derived growth factor receptors: determination of a pharmacokinetic/pharmacodynamic relationship. Clinical cancer research. 2003, 9 (1), 327–337. [PubMed] [Google Scholar]
- Chowdhary S.; Shalini; Arora A.; Kumar V. A mini review on isatin, an anticancer scaffold with potential activities against neglected tropical diseases (NTDs). Pharmaceuticals. 2022, 15 (5), 536. 10.3390/ph15050536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prenen H.; Cools J.; Mentens N.; et al. Efficacy of the kinase inhibitor SU11248 against gastrointestinal stromal tumor mutants refractory to imatinib mesylate. Clin. Cancer Res. 2006, 12 (8), 2622–2627. 10.1158/1078-0432.CCR-05-2275. [DOI] [PubMed] [Google Scholar]
- Siegel R. L.; Miller K. D.; Wagle N. S.; Jemal A. Cancer statistics, 2023. Ca Cancer J. Clin. 2023, 73 (1), 17–48. 10.3322/caac.21763. [DOI] [PubMed] [Google Scholar]
- Balogh J.; Victor D.; Asham E. H; Burroughs S. G.; Boktour M.; Saharia A.; Li X.; Ghobrial M.; Monsour H. Hepatocellular carcinoma: a review. Journal of hepatocellular carcinoma. 2016, 3, 41–53. 10.2147/JHC.S61146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruix J.; Gores G. J.; Mazzaferro V. Hepatocellular carcinoma: clinical frontiers and perspectives. Gut. 2014, 63 (5), 844–855. 10.1136/gutjnl-2013-306627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorgeirsson S. S.; Grisham J. W. Molecular pathogenesis of human hepatocellular carcinoma. Nature genetics. 2002, 31 (4), 339–346. 10.1038/ng0802-339. [DOI] [PubMed] [Google Scholar]
- Shire A.; Roberts L. Prevention of hepatocellular carcinoma: progress and challenges. Minerva gastroenterologica e dietologica. 2012, 58 (1), 49–64. [PubMed] [Google Scholar]
- Aneja B.; Khan N. S.; Khan P.; et al. Design and development of Isatin-triazole hydrazones as potential inhibitors of microtubule affinity-regulating kinase 4 for the therapeutic management of cell proliferation and metastasis. Eur. J. Med. Chem. 2019, 163, 840–852. 10.1016/j.ejmech.2018.12.026. [DOI] [PubMed] [Google Scholar]
- Ahmed S.; Khan P.; Irfan I.; et al. Structure-guided design and development of vanillin-triazole conjugates as potential MARK4 inhibitors targeting hepatocellular carcinoma. J. Mol. Struct. 2023, 1293, 136303. 10.1016/j.molstruc.2023.136303. [DOI] [Google Scholar]
- Maestro, Version 9.0; Schrödinger: New York, 2009. [Google Scholar]
- Jacobson M. P.; Pincus D. L.; Rapp C. S.; et al. A hierarchical approach to all-atom protein loop prediction. Proteins: Structure, Function, and Bioinformatics. 2004, 55 (2), 351–367. 10.1002/prot.10613. [DOI] [PubMed] [Google Scholar]
- Madhavi Sastry G.; Adzhigirey M.; Day T.; Annabhimoju R.; Sherman W. Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. Journal of computer-aided molecular design. 2013, 27, 221–234. 10.1007/s10822-013-9644-8. [DOI] [PubMed] [Google Scholar]
- Li J.; Abel R.; Zhu K.; Cao Y.; Zhao S.; Friesner R. A. The VSGB 2.0 model: a next generation energy model for high resolution protein structure modeling. Proteins: Structure, Function, and Bioinformatics. 2011, 79 (10), 2794–2812. 10.1002/prot.23106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos K.; Wu C.; Damm W.; et al. OPLS3e: Extending force field coverage for drug-like small molecules. Journal of chemical theory and computation. 2019, 15 (3), 1863–1874. 10.1021/acs.jctc.8b01026. [DOI] [PubMed] [Google Scholar]
- Friesner R. A.; Banks J. L.; Murphy R. B.; et al. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. Journal of medicinal chemistry. 2004, 47 (7), 1739–1749. 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- Friesner R. A.; Murphy R. B.; Repasky M. P.; et al. Extra precision glide: Docking and scoring incorporating a model of hydrophobic enclosure for protein- ligand complexes. Journal of medicinal chemistry. 2006, 49 (21), 6177–6196. 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- Halgren T. A.; Murphy R. B.; Friesner R. A.; et al. Glide: a new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. Journal of medicinal chemistry. 2004, 47 (7), 1750–1759. 10.1021/jm030644s. [DOI] [PubMed] [Google Scholar]
- Jorgensen W. L.; Madura J. D. Temperature and size dependence for Monte Carlo simulations of TIP4P water. Mol. Phys. 1985, 56 (6), 1381–1392. 10.1080/00268978500103111. [DOI] [Google Scholar]
- Lawrence C.; Skinner J. Flexible TIP4P model for molecular dynamics simulation of liquid water. Chemical physics letters. 2003, 372 (5–6), 842–847. 10.1016/S0009-2614(03)00526-8. [DOI] [Google Scholar]
- Martyna G. J.; Tobias D. J.; Klein M. L. Constant pressure molecular dynamics algorithms. Journal of chemical physics. 1994, 101 (5), 4177–4189. 10.1063/1.467468. [DOI] [Google Scholar]
- Martyna G. J.; Klein M. L.; Tuckerman M. Nosé-Hoover chains: The canonical ensemble via continuous dynamics. Journal of chemical physics. 1992, 97 (4), 2635–2643. 10.1063/1.463940. [DOI] [Google Scholar]
- Martyna G. J.; Tuckerman M. E.; Tobias D. J.; Klein M. L. Explicit reversible integrators for extended systems dynamics. Mol. Phys. 1996, 87 (5), 1117–1157. 10.1080/00268979600100761. [DOI] [Google Scholar]
- Naz F.; Sami N.; Islam A.; Ahmad F.; Hassan M. I. Ubiquitin-associated domain of MARK4 provides stability at physiological pH. International journal of biological macromolecules. 2016, 93, 1147–1154. 10.1016/j.ijbiomac.2016.09.087. [DOI] [PubMed] [Google Scholar]
- Aneja B.; Khan N. S.; Khan P.; et al. Design and development of Isatin-triazole hydrazones as potential inhibitors of microtubule affinity-regulating kinase 4 for the therapeutic management of cell proliferation and metastasis. Eur. J. Med. Chem. 2019, 163, 840–852. 10.1016/j.ejmech.2018.12.026. [DOI] [PubMed] [Google Scholar]
- Yousuf M.; Shamsi A.; Khan P.; et al. Ellagic acid controls cell proliferation and induces apoptosis in breast cancer cells via inhibition of cyclin-dependent kinase 6. International journal of molecular sciences. 2020, 21 (10), 3526. 10.3390/ijms21103526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naz H.; Khan P.; Tarique M.; et al. Binding studies and biological evaluation of β-carotene as a potential inhibitor of human calcium/calmodulin-dependent protein kinase IV. International journal of biological macromolecules. 2017, 96, 161–170. 10.1016/j.ijbiomac.2016.12.024. [DOI] [PubMed] [Google Scholar]
- Das Mahapatra A.; Queen A.; Yousuf M.; Khan P.; Hussain A.; Rehman M. T.; Alajmi M. F.; Datta B.; Hassan M. I. Design and development of 5-(4H)-oxazolones as potential inhibitors of human carbonic anhydrase VA: towards therapeutic management of diabetes and obesity. Journal of Biomolecular Structure and Dynamics. 2022, 40 (7), 3144–3154. 10.1080/07391102.2020.1845803. [DOI] [PubMed] [Google Scholar]
- Khan P.; Queen A.; Mohammad T.; et al. Identification of α-mangostin as a potential inhibitor of microtubule affinity regulating kinase 4. Journal of natural products. 2019, 82 (8), 2252–2261. 10.1021/acs.jnatprod.9b00372. [DOI] [PubMed] [Google Scholar]
- Aneja B.; Queen A.; Khan P.; et al. Design, synthesis & biological evaluation of ferulic acid-based small molecule inhibitors against tumor-associated carbonic anhydrase IX. Bioorg. Med. Chem. 2020, 28 (9), 115424. 10.1016/j.bmc.2020.115424. [DOI] [PubMed] [Google Scholar]
- Aneja B.; Queen A.; Khan P.; et al. Design, synthesis & biological evaluation of ferulic acid-based small molecule inhibitors against tumor-associated carbonic anhydrase IX. Bioorg. Med. Chem. 2020, 28 (9), 115424. 10.1016/j.bmc.2020.115424. [DOI] [PubMed] [Google Scholar]
- Shamsi F.; Hasan P.; Queen A.; et al. Synthesis and SAR studies of novel 1,2,4-oxadiazole-sulfonamide based compounds as potential anticancer agents for colorectal cancer therapy. Bioorg Chem. 2020, 98, 103754. 10.1016/j.bioorg.2020.103754. [DOI] [PubMed] [Google Scholar]
- Mishra P.; Mishra A.; Bahe A. K.; Roy A.; Das R. Synthesis of Isatin and its derivatives containing heterocyclic compounds. Journal of the Turkish Chemical Society Section A: Chemistry. 2021, 8 (4), 1089–1098. 10.18596/jotcsa.962260. [DOI] [Google Scholar]
- Irfan I.; Ali A.; Reddi B.; et al. Design, Synthesis and Mechanistic Studies of Novel Isatin-Pyrazole Hydrazone Conjugates as Selective and Potent Bacterial MetAP Inhibitors. Antibiotics. 2022, 11 (8), 1126. 10.3390/antibiotics11081126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naz F.; Anjum F.; Islam A.; Ahmad F.; Hassan M. I. Microtubule affinity-regulating kinase 4: structure, function, and regulation. Cell biochemistry and biophysics. 2013, 67, 485–499. 10.1007/s12013-013-9550-7. [DOI] [PubMed] [Google Scholar]
- Khan P.; Idrees D.; Moxley M. A.; Corbett J. A.; Ahmad F.; von Figura G.; Sly W. S.; Waheed A.; Hassan M. I. Hassan MI Luminol Based Chemiluminescent Signals: Clinical and Non-Clinical Application and Future Uses. Appl. Biochem. Biotechnol. 2014, 173 (2), 333–355. 10.1007/s12010-014-0850-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed S.; Mobashir M.; Al-Keridis L. A.; Alshammari N.; Adnan M.; Abid M.; Hassan M. I. A network-guided approach to discover phytochemical-based anticancer therapy targeting MARK4 for hepatocellular carcinoma. Frontiers in Oncology. 2022, 12, 3379. 10.3389/fonc.2022.914032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad T.; Khan F. I.; Lobb K. A.; Islam A.; Ahmad F.; Hassan M. I. Identification and evaluation of bioactive natural products as potential inhibitors of human microtubule affinity-regulating kinase 4 (MARK4). Journal of Biomolecular Structure and Dynamics. 2019, 37 (7), 1813–1829. 10.1080/07391102.2018.1468282. [DOI] [PubMed] [Google Scholar]
- Natarajan S. R.; Ponnusamy L.; Manoharan R. MARK2/4 promotes Warburg effect and cell growth in non-small cell lung carcinoma through the AMPKα1/mTOR/HIF-1α signaling pathway. Biochimica et Biophysica Acta (BBA)-Molecular Cell Research. 2022, 1869 (7), 119242. 10.1016/j.bbamcr.2022.119242. [DOI] [PubMed] [Google Scholar]
- Flores-Rodriguez P.; Harrington C. R.; Wischik C. M.; Ibarra-Bracamontes V.; Zarco N.; Navarrete A.; Martinez-Maldonado A.; Guadarrama-Ortiz P.; Villanueva-Fierro I.; Ontiveros-Torres M. A.; et al. Phospho-tau protein expression in the cell cycle of SH-SY5Y neuroblastoma cells: a morphological study. Journal of Alzheimer’s Disease. 2019, 71 (2), 631–645. 10.3233/JAD-190155. [DOI] [PubMed] [Google Scholar]
- Eldeeb M.; Sanad E. F.; Ragab A.; et al. Anticancer effects with molecular docking confirmation of newly synthesized isatin sulfonamide molecular hybrid derivatives against hepatic cancer cell lines. Biomedicines. 2022, 10 (3), 722. 10.3390/biomedicines10030722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eldehna W. M.; Almahli H.; Al-Ansary G. H.; et al. Synthesis and in vitro anti-proliferative activity of some novel isatins conjugated with quinazoline/phthalazine hydrazines against triple-negative breast cancer MDA-MB-231 cells as apoptosis-inducing agents. Journal of Enzyme Inhibition and Medicinal Chemistry. 2017, 32 (1), 600–613. 10.1080/14756366.2017.1279155. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data analyzed during this study are included in this manuscript and the Supporting Information attached to this paper.