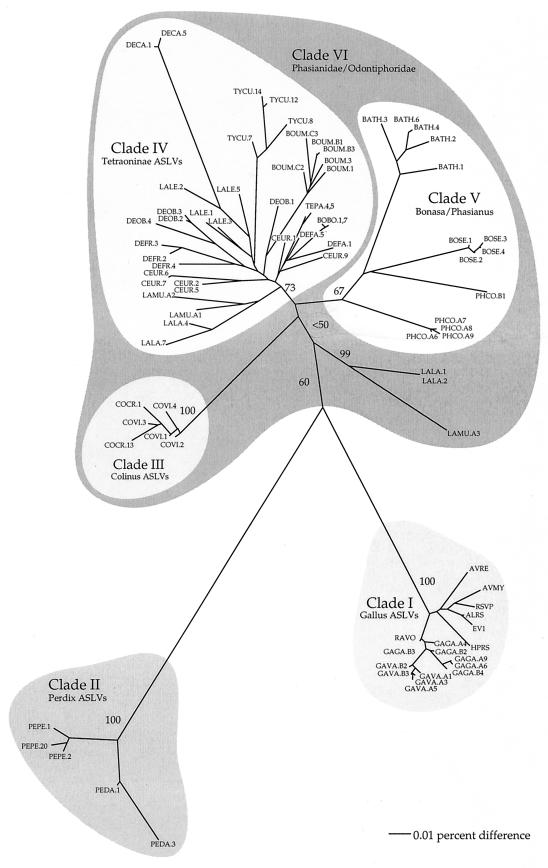
FIG. 2.
Unrooted NJ tree constructed using Kimura's two-parameter corrected distances, showing the relationship of 85 retroviral sequences based on 720 nucleotide sites from fragment 1 (Fig. 1A) of the gag gene. Bootstrap values are presented for the earliest divergences. An NJ tree using all 112 retroviral sequences has essentially the same topology; we removed 27 taxa representing multiple clones from single individuals from the analysis shown for clarity. Viral sequences are named for their host species (see Table 1 for abbreviations) and a number that identifies clones from the same host individual. When clones were sequenced from multiple individuals from a single species, a letter identifies the individual.