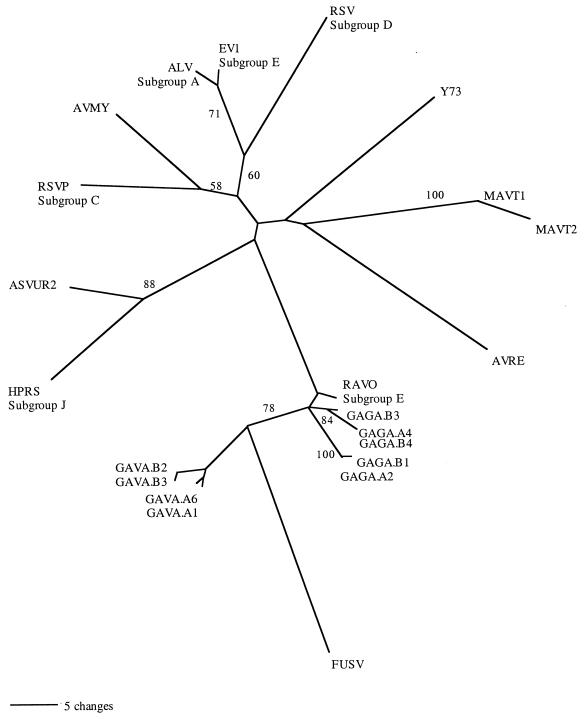
FIG. 5.
Unrooted MP analysis of ASLV gag DNA sequences isolated from birds in the genus Gallus. This phylogeny is based on 1,165 characters (fragment 2) of which 95 are parsimony informative. This is one of two equally parsimonious hypotheses. Bootstrap values above 50 are shown on branches (100 replicates). ML analysis using the GTR model yields the same topology. Sequences GAGA and GAVA are newly sequenced elements from two individuals (A and B) of Gallus gallus and Gallus varius, respectively. gag sequences for the following viruses were obtained from databases: avian leukemia virus, subgroup A (ALV); exogenous avian leukosis virus, subgroup J (HPRS-103); avian myelocytomatosis virus (AVMY); avian retrovirus IC10 (AVRE); RSV (Prague), subgroup C (RSVP); RSV (Schmidt-Ruppin), subgroup D (RSV); FUSV; myeloblastosis-associated virus 1 (MAVT1); myeloblastosis-associated virus 2 (MAVT2); avian sarcoma virus Y73 (Y73); avian sarcoma virus UR2 (ASVUR2); Rous sarcoma-defective endogenous virus, subgroup E (EV1); chicken provirus RAV-0, subgroup E (RAV0) (accession numbers are listed in Materials and Methods).