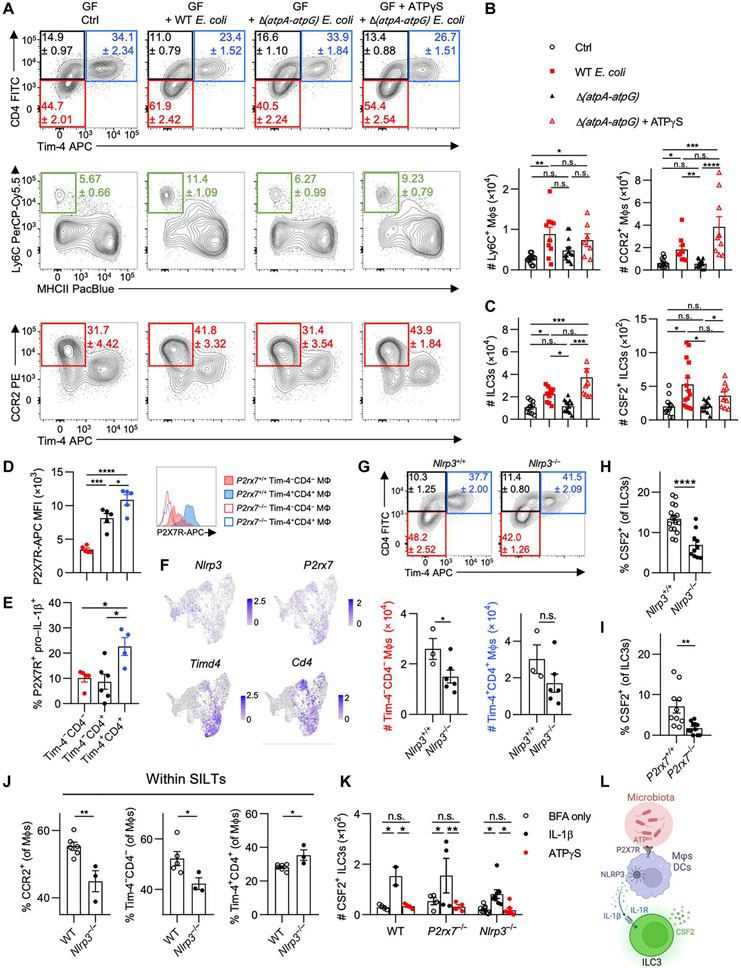
Fig. 4. Microbial ATP regulates MΦ composition and drives CSF2 production by ILC3s through P2X7R and NLRP3 inflammasome signaling.
(A to C) GF mice were orally gavaged with WT or Δ(atpA-atpG) Strr E. coli MG1655, with a group of Δ(atpA-atpG) Strr E. coli MG1655 recipients receiving daily intrarectal applications of ATPγS. Mice were analyzed 1 week later. (A) Representative flow cytometry plots show colonic monocyte and MΦ composition. (B) Quantifications of Ly6C+ and CCR2+ MΦs. (C) Quantifications of total ILC3s and CSF2-producing ILC3s in the colon. (D and E) Each WT colonic MΦ population was assessed for (D) median fluorescence intensity (MFI) of P2X7R staining, with histograms (right) showing staining in P2rx7+/+ versus P2rx7−/− mice, or (E) percentage of P2X7R+ pro–IL-1β+ cells. (F) Feature plots illustrating expression of indicated genes in colonic MΦs/monocytes. (G) Contour plots (top) and quantification (bottom) of colonic MΦs in B6 and Nlrp3−/− littermates. (H and I) Percentage of CSF2-producing colonic ILC3s in Nlrp3−/− and P2rx7−/− mice with respective littermate controls. (J) WT and Nlrp3−/− colonic SILTs were isolated and assessed for percentage of each MΦ population. (K) Colonic LP leukocytes were treated with vehicle control, IL-1β, or ATPγS in the presence of brefeldin A (BFA) and quantified for CSF2+ ILC3s. (L) Schematic overview of how microbiota-derived ATPex regulates ILC3-derived CSF2. For (J), each data point represents pooled SILTs from two or three mice. One-way ANOVA with post hoc Tukey’s test (B to E), two-way ANOVA with post hoc Šidák’s multiple comparisons test (K), or unpaired Student’s t test (G to J) was performed.